Matt Doran
@matthdoran.bsky.social
130 followers
230 following
8 posts
Structural biologist and post-doc in the Brown Lab @harvardmed. Formerly at the Lehman lab @BUMedicine.
Posts
Media
Videos
Starter Packs
Reposted by Matt Doran
Yi-Wei Chang
@yiweichang.bsky.social
· May 15
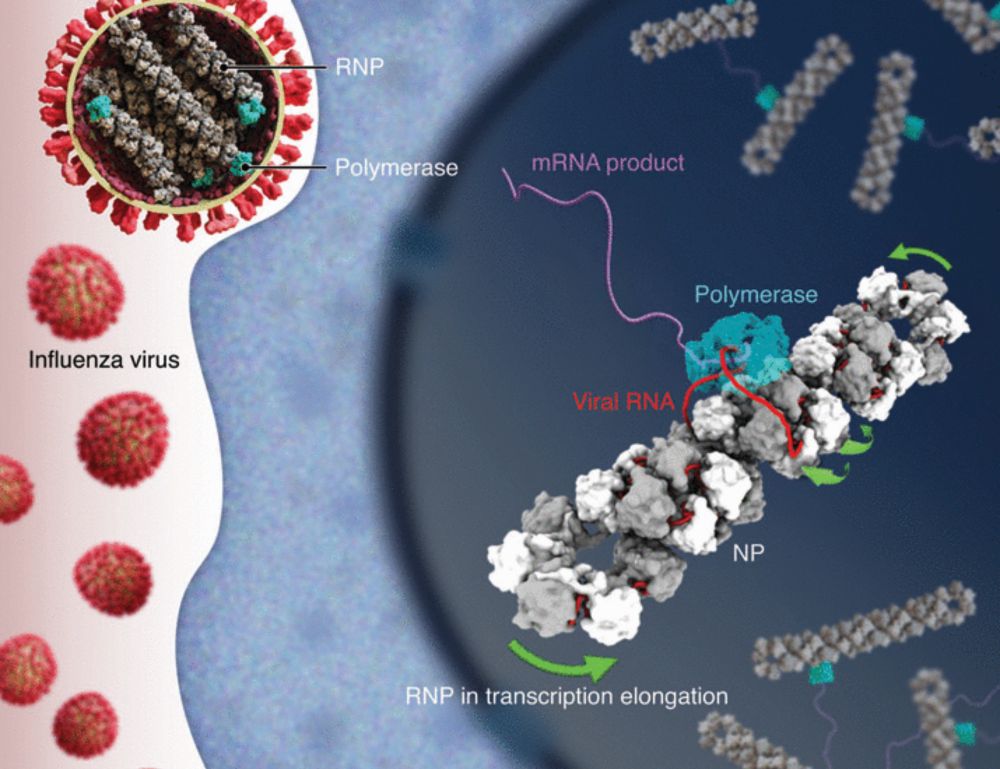
Molecular basis of influenza ribonucleoprotein complex assembly and processive RNA synthesis
Influenza viruses replicate and transcribe their genome in the context of a conserved ribonucleoprotein (RNP) complex. By integrating cryo–electron microscopy single-particle analysis and cryo–electro...
www.science.org
Reposted by Matt Doran
Sven Lange
@sven-m-lange.bsky.social
· Apr 29

A conserved mechanism for the retrieval of polyubiquitinated proteins from cilia
The temporospatial distribution of proteins within cilia is regulated by intraflagellar transport (IFT), wherein molecular trains shuttle between the cell body and cilium. Defects in this process impair various signal-transduction pathways and cause ciliopathies. Although K63-linked ubiquitination appears to trigger protein export from cilia, the mechanisms coupling polyubiquitinated proteins to IFT remain unclear. Using a multidisciplinary approach, we demonstrate that a complex of CFAP36, a conserved ciliary protein of previously unknown function, and ARL3, a GTPase involved in ciliary import, binds polyubiquitinated proteins and links them to retrograde IFT trains. CFAP36 uses a coincidence detection mechanism to simultaneously bind two IFT subunits accessible only in retrograde trains. Depleting CFAP36 accumulates K63-linked ubiquitin in cilia and disrupts Hedgehog signaling, a pathway reliant on the retrieval of ubiquitinated receptors. These findings advance our understanding of ubiquitin-mediated protein transport and ciliary homeostasis, and demonstrate how structural changes in IFT trains achieve cargo selectivity. ### Competing Interest Statement The authors have declared no competing interest. Sara Elizabeth O'Brien Trust Postdoctoral Fellowship awarded through the Charles A. King Trust Postdoctoral Research Fellowship Program, , 8460873-01 Richard and Susan Smith Family Foundation, https://ror.org/05j95n956, National Institute of General Medical Sciences (NIGMS), , R01GM141109, R01GM143183
www.biorxiv.org
Matt Doran
@matthdoran.bsky.social
· Mar 13
Matt Doran
@matthdoran.bsky.social
· Mar 13
Matt Doran
@matthdoran.bsky.social
· Mar 13
Matt Doran
@matthdoran.bsky.social
· Mar 13
Matt Doran
@matthdoran.bsky.social
· Mar 13
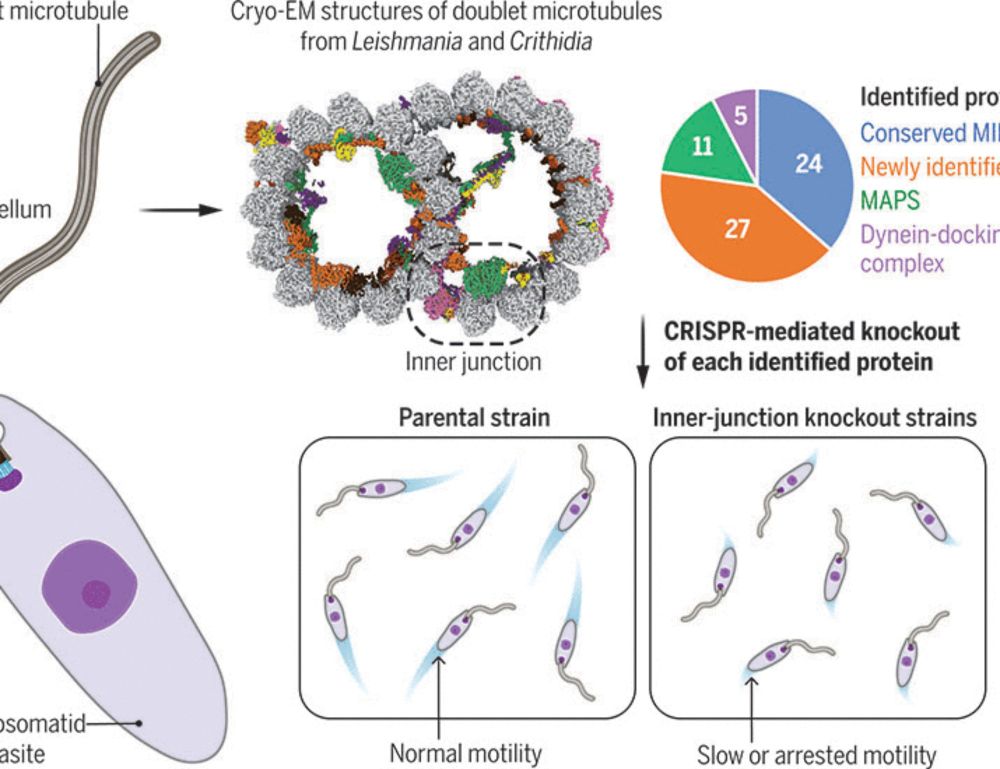
Evolutionary adaptations of doublet microtubules in trypanosomatid parasites
The movement and pathogenicity of trypanosomatid species, the causative agents of trypanosomiasis and leishmaniasis, are dependent on a flagellum that contains an axoneme of dynein-bound doublet micro...
tinyurl.com


