Comp & Exp Biochemist, Protein Engineer, 'Would-be designer' (F. Arnold) | SynBio | HT Screens & Selections | Nucleic Acid Enzymes | Biocatalysis | Rstats & Datavis
https://www.fuerstlab.com
https://orcid.org/0000-0001-7720-9
16/19

16/19
Without a PDB to compare to, you can also rescue via a simple trick: just compute statistical outliers from an initial alignment, realign without them, and use median RMSD
15/19

Without a PDB to compare to, you can also rescue via a simple trick: just compute statistical outliers from an initial alignment, realign without them, and use median RMSD
15/19
14/19

14/19
13/19

13/19
11/19

11/19
Duh: empty MSA == ss mode
9/19

Duh: empty MSA == ss mode
9/19
8/19

8/19
As others noted: the signal from these MSAs / pLM embeddings overrules “reason”
7/19

As others noted: the signal from these MSAs / pLM embeddings overrules “reason”
7/19
First, we checked how good they are at identifying clearly bad designs
5/19

First, we checked how good they are at identifying clearly bad designs
5/19
2/19

2/19
Imagine (re)designing a protein via inverse folding. AF2 predicts the designed sequence to a structure with pLDDT 94 & you get 1.8 Å RMSD to the input. Perfect design?
What if I told u that the structure has 4 solvent-exposed Trp and 3 Pro where a Gly should be?
Why to be wary🧵👇
Imagine (re)designing a protein via inverse folding. AF2 predicts the designed sequence to a structure with pLDDT 94 & you get 1.8 Å RMSD to the input. Perfect design?
What if I told u that the structure has 4 solvent-exposed Trp and 3 Pro where a Gly should be?
Why to be wary🧵👇
Here search results from on "DNA", last year (orange) vs now (blue) and the indexing issue is only noticeable for the current year ('24), barely an increase for '23
New hypothesis: all (bio?) topics peaked around '21?

Here search results from on "DNA", last year (orange) vs now (blue) and the indexing issue is only noticeable for the current year ('24), barely an increase for '23
New hypothesis: all (bio?) topics peaked around '21?
Too lazy to extract the numbers and make a scatter plot, but I think Ala and Glu are at least as off the axis as the aromatics
www.uniprot.org/uniprotkb/st...

Too lazy to extract the numbers and make a scatter plot, but I think Ala and Glu are at least as off the axis as the aromatics
www.uniprot.org/uniprotkb/st...
Here formatted in a similar way from source www.bka.de/DE/AktuelleI...

Here formatted in a similar way from source www.bka.de/DE/AktuelleI...
fuerstlab.shinyapps.io/SeqNovelty/
quick 🧵
fuerstlab.shinyapps.io/SeqNovelty/
quick 🧵
Original (very awkward) color scale did not reflect how much of an outlier NL is.
Picture changes for 3yr - school age though.
Of course this data does not distinguish between day care vs grandparents etc. Not that the latter is an option for expats, anyway 😶🌫️


Original (very awkward) color scale did not reflect how much of an outlier NL is.
Picture changes for 3yr - school age though.
Of course this data does not distinguish between day care vs grandparents etc. Not that the latter is an option for expats, anyway 😶🌫️



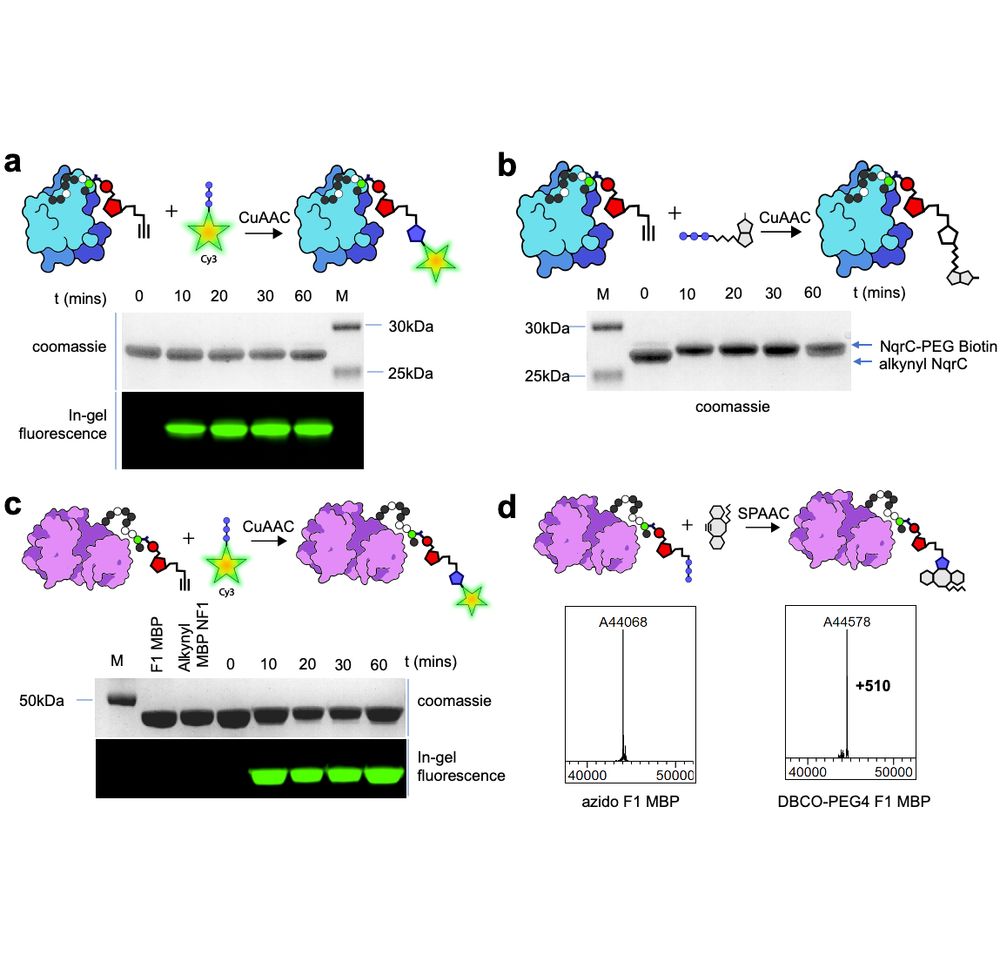
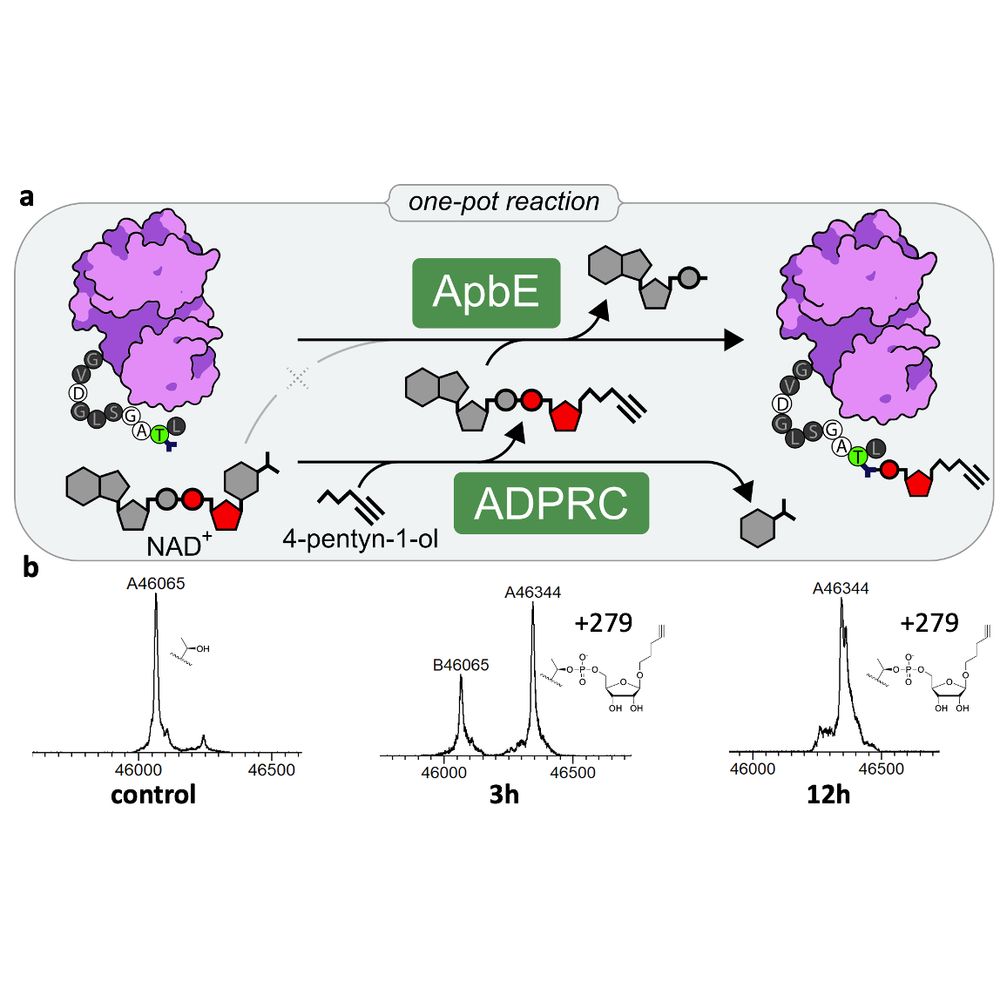
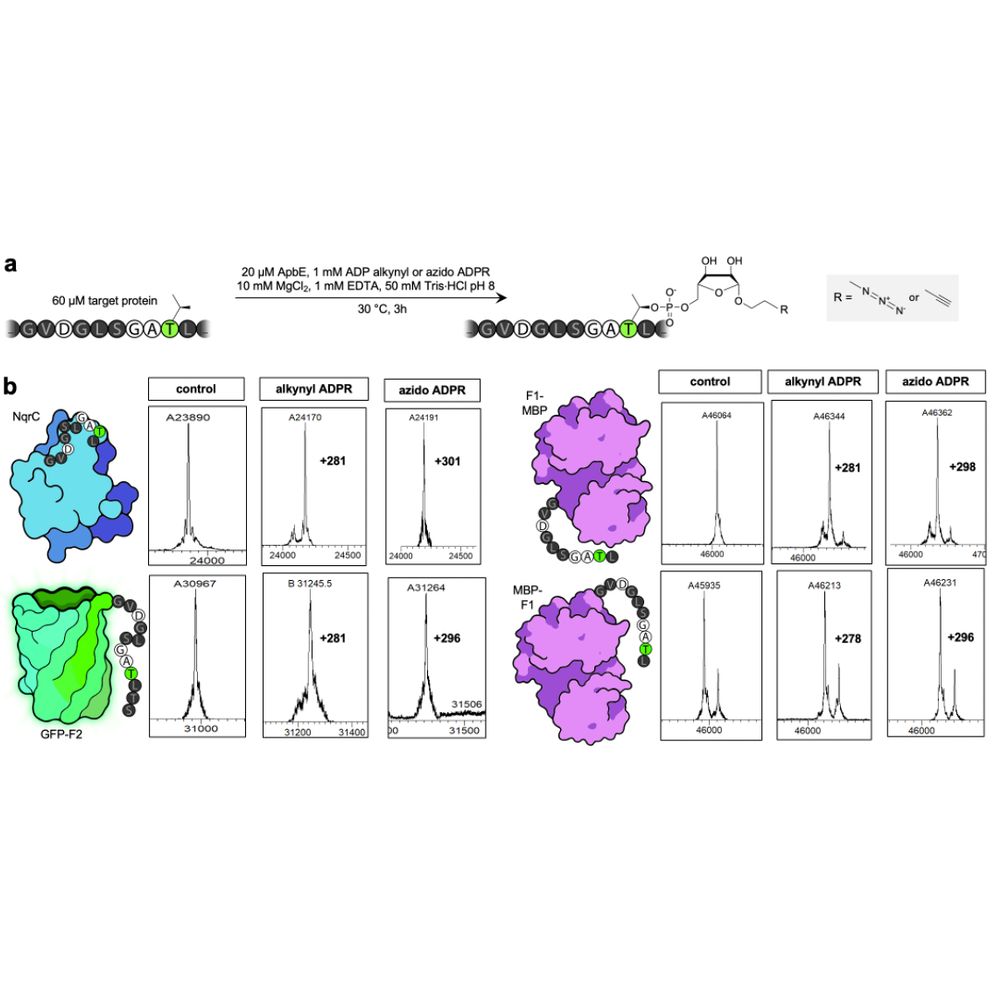
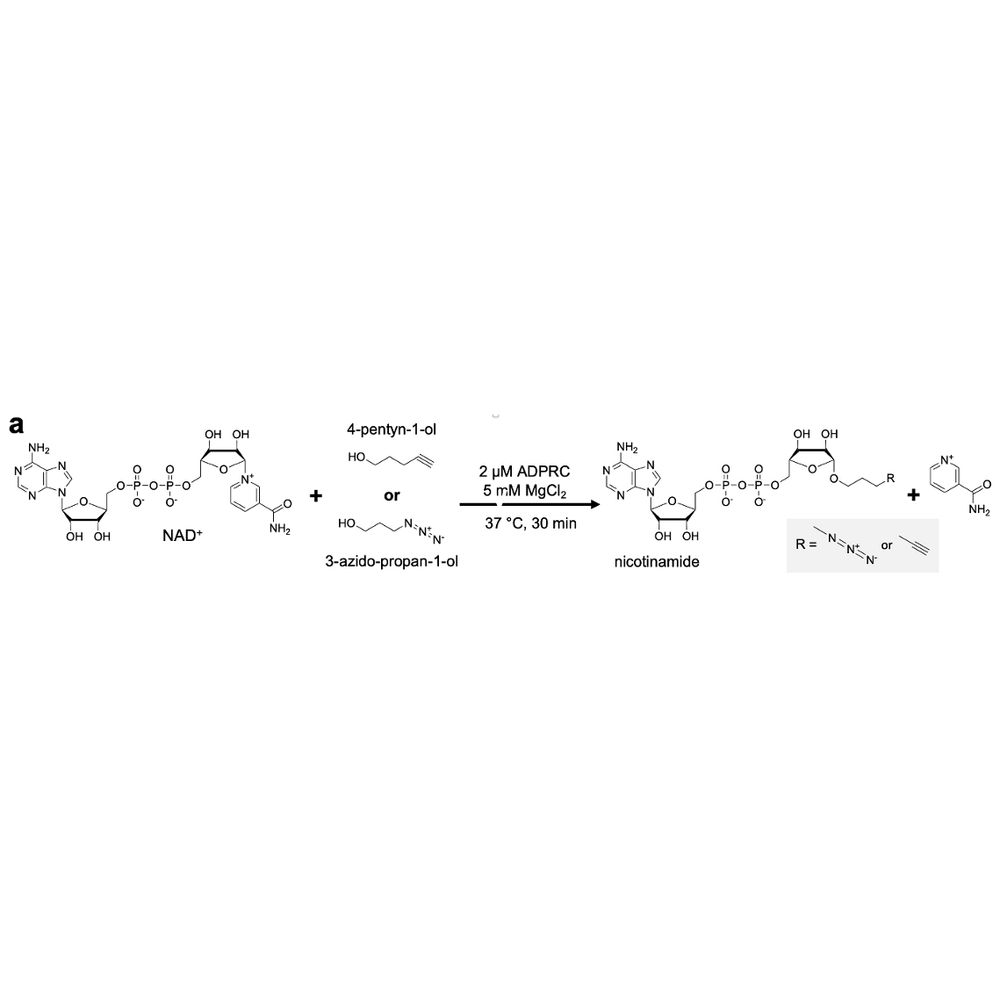
We prouldy present: ADD-tagging of proteins (or "ADDing") —a super convenient enzymatic technique to install click chemistry handles on proteins.
Led by superstar @wahyuwidodo.bsky.social
www.biorxiv.org/content/10.1...
A 🧵👇🏽
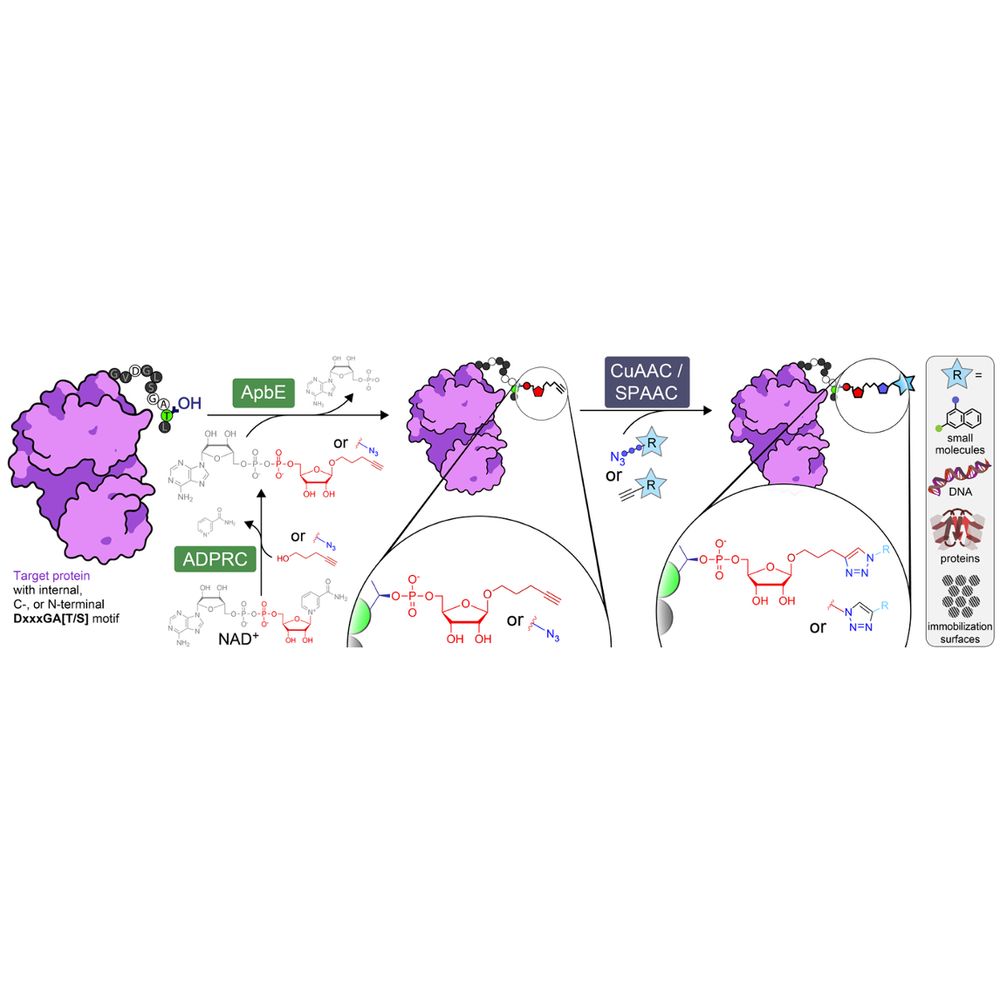
We prouldy present: ADD-tagging of proteins (or "ADDing") —a super convenient enzymatic technique to install click chemistry handles on proteins.
Led by superstar @wahyuwidodo.bsky.social
www.biorxiv.org/content/10.1...
A 🧵👇🏽

