
Then, we identified TE silencing factors and found an expansion of piwil1 genes in Lake Malawi cichlids, which was likely driven...
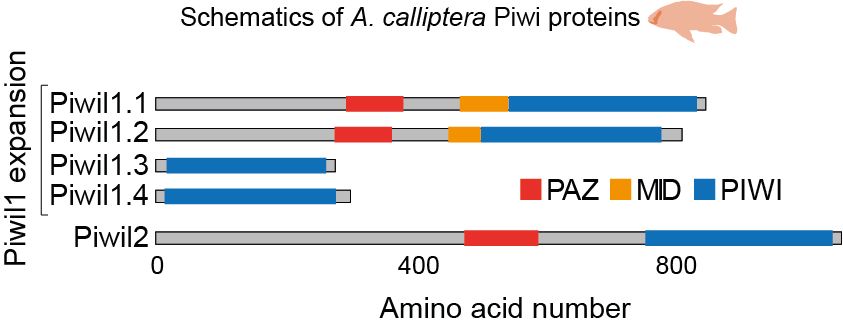
Then, we identified TE silencing factors and found an expansion of piwil1 genes in Lake Malawi cichlids, which was likely driven...


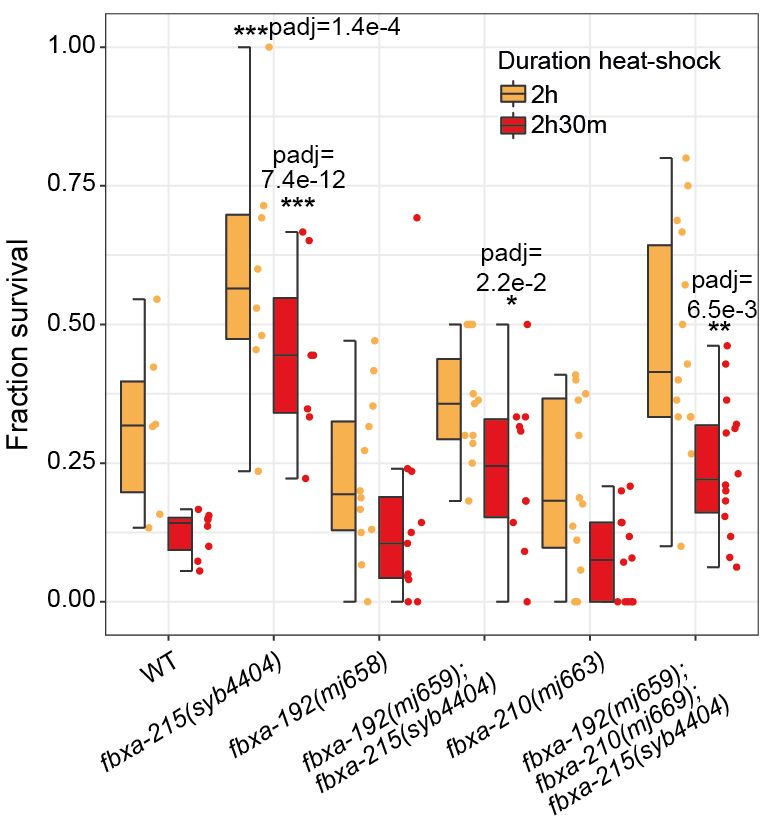
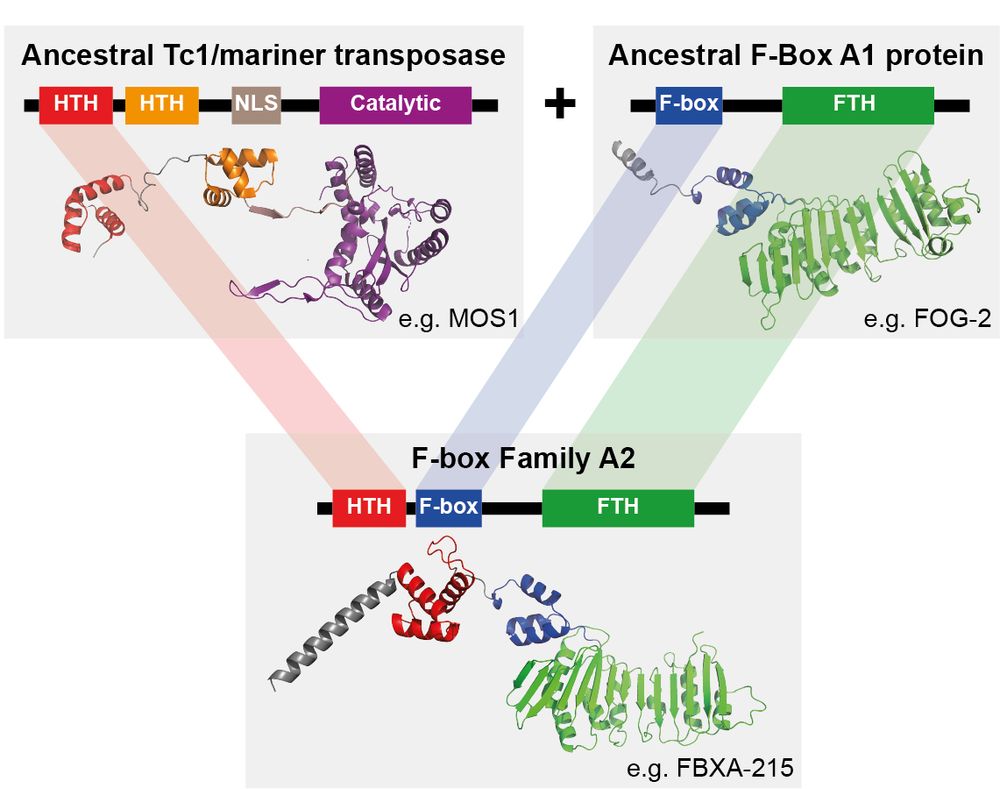
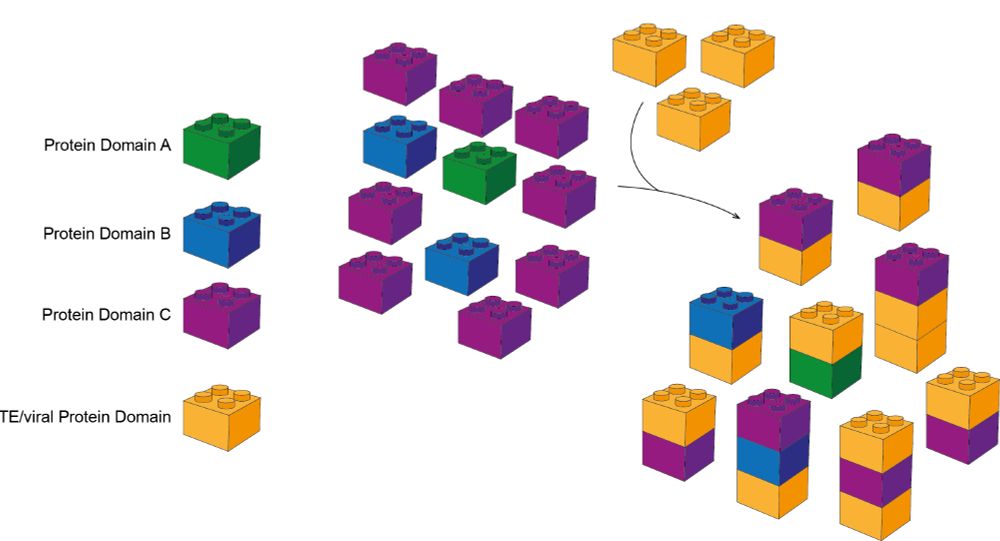
Short 🧵 with highlights.

Short 🧵 with highlights.

