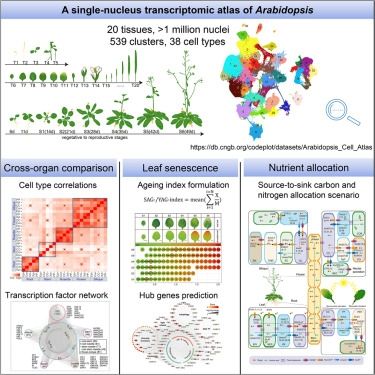
www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...
Our approach allows silencing defense systems of choice. We show how this approach enables programming of “untransformable” bacteria, and how it can enhance phage therapy applications
Congrats Jeremy Garb!
tinyurl.com/Syttt
🧵

Our approach allows silencing defense systems of choice. We show how this approach enables programming of “untransformable” bacteria, and how it can enhance phage therapy applications
Congrats Jeremy Garb!
tinyurl.com/Syttt
🧵
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
How broad is their Pangenome?
How will you choose representatives from the whole collection?
Look no further! PanGene-O-Meter is here for you!
From @haimashkenazy.bsky.social @plantevolution.bsky.social
www.biorxiv.org/content/10.1...

How broad is their Pangenome?
How will you choose representatives from the whole collection?
Look no further! PanGene-O-Meter is here for you!
From @haimashkenazy.bsky.social @plantevolution.bsky.social
www.biorxiv.org/content/10.1...
Several groups have presented sc/snRNA-seq analyses of Arabidopsis leaves after bacterial infection before. Is there anything left to discover? Turns out: Yes.
#plantscience
www.biorxiv.org/content/10.1...
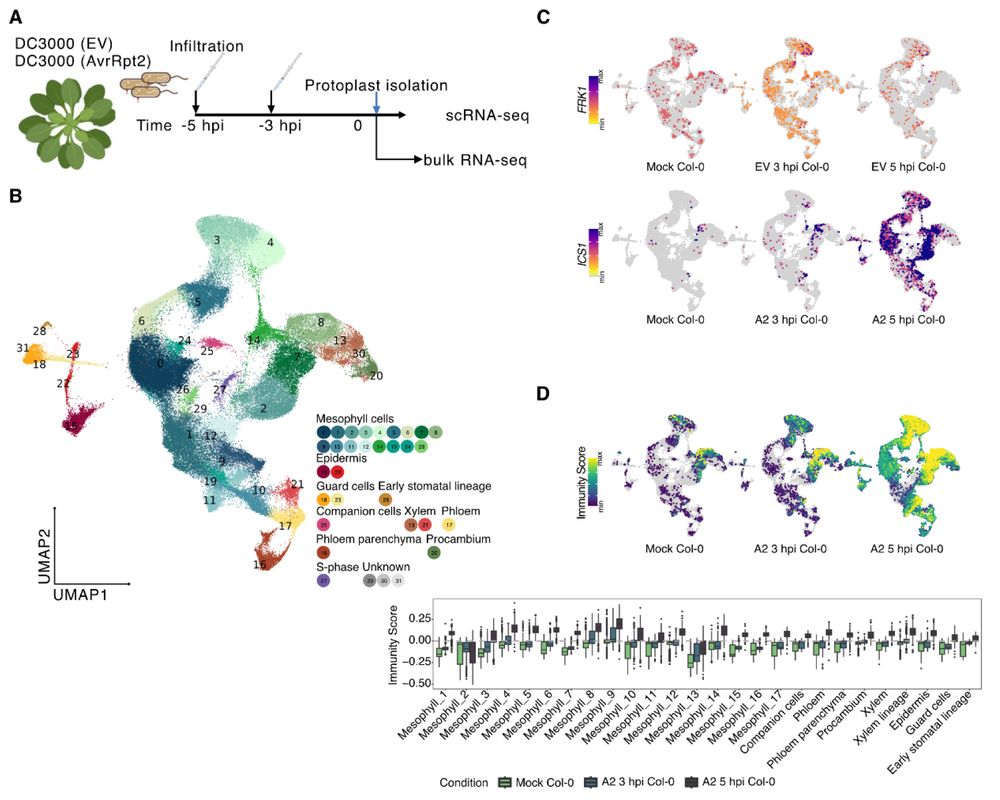
Several groups have presented sc/snRNA-seq analyses of Arabidopsis leaves after bacterial infection before. Is there anything left to discover? Turns out: Yes.
#plantscience
www.biorxiv.org/content/10.1...
#plantscience
link.springer.com/article/10.1...?
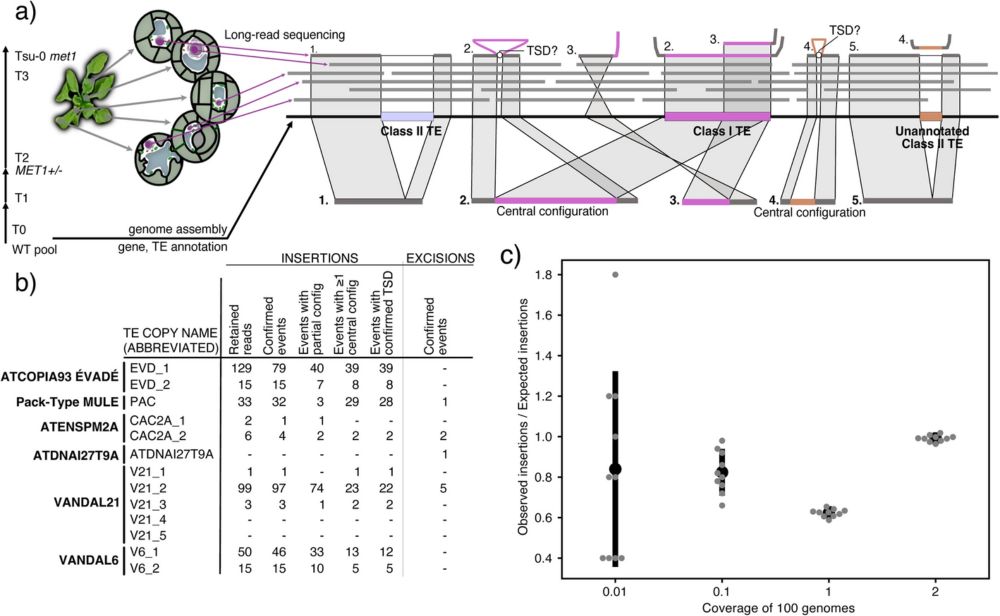
#plantscience
link.springer.com/article/10.1...?
Our AlphaFold sensor vs helper NLR paper is now published in @newphyt.bsky.social!
Big thanks to the reviewers for their constructive feedback 🙏
📖 Check out the peer-reviewed version here 👇
🔗 nph.onlinelibrary.wiley.com/doi/10.1111/...
bsky.app/profile/yusu...
Our AlphaFold sensor vs helper NLR paper is now published in @newphyt.bsky.social!
Big thanks to the reviewers for their constructive feedback 🙏
📖 Check out the peer-reviewed version here 👇
🔗 nph.onlinelibrary.wiley.com/doi/10.1111/...
bsky.app/profile/yusu...

by Chandler Sutherland @danimstevens.bsky.social @weiweibio.bsky.social Kyungyong Seong
academic.oup.com/plcell/advan...

by Chandler Sutherland @danimstevens.bsky.social @weiweibio.bsky.social Kyungyong Seong
academic.oup.com/plcell/advan...

🧑🔬 Yanhao Cheng, Yiping Qi, et al.
📔 @theplantcell.bsky.social
🔗 academic.oup.com/plcell/advan...
#️⃣ #PlantScience #PlantCRISPR #PlantMethods #PlantGenomics #PlantNGTs #PlantGE #PlantGenomeEditing
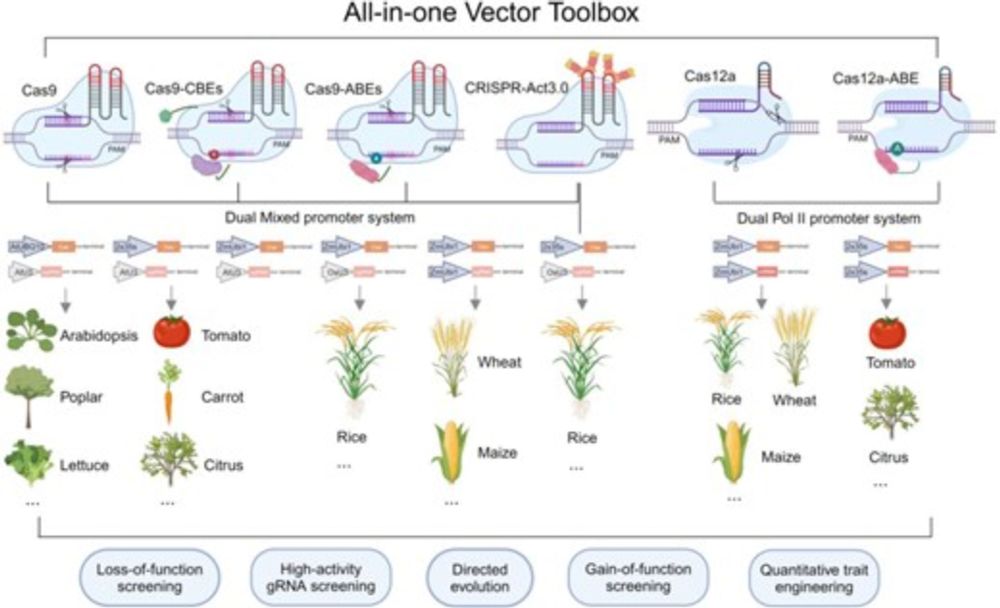
🧑🔬 Yanhao Cheng, Yiping Qi, et al.
📔 @theplantcell.bsky.social
🔗 academic.oup.com/plcell/advan...
#️⃣ #PlantScience #PlantCRISPR #PlantMethods #PlantGenomics #PlantNGTs #PlantGE #PlantGenomeEditing
#plantscience
www.biorxiv.org/content/10.1...
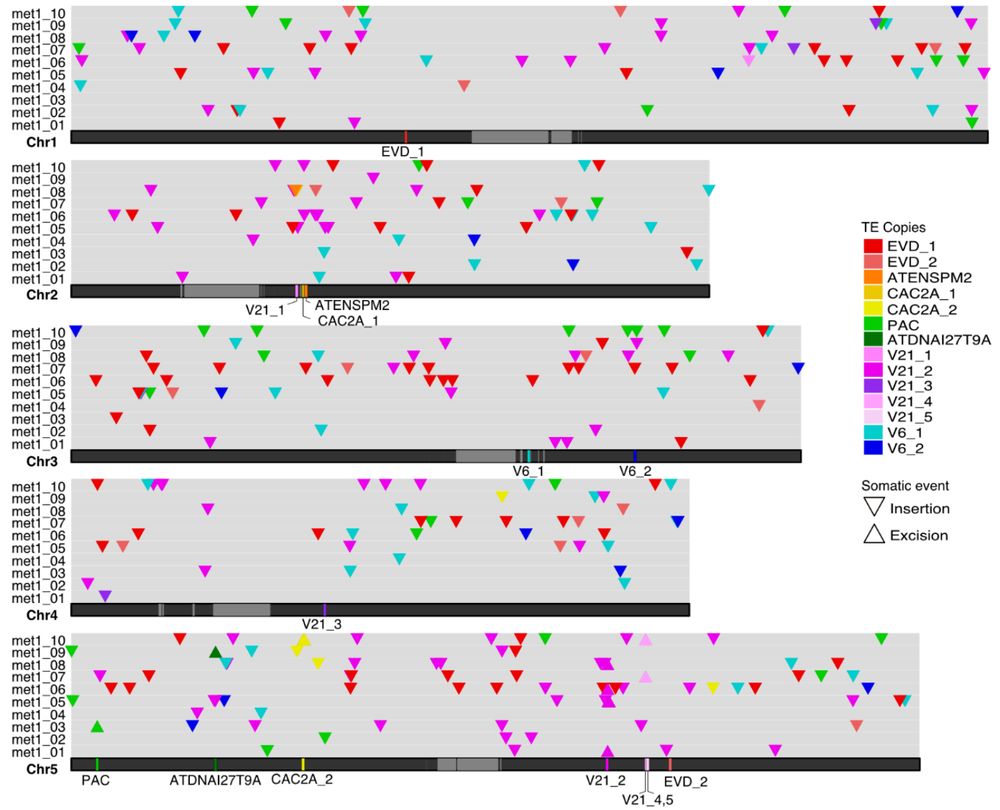
#plantscience
www.biorxiv.org/content/10.1...

#PlantScience
www.nature.com/articles/s41...
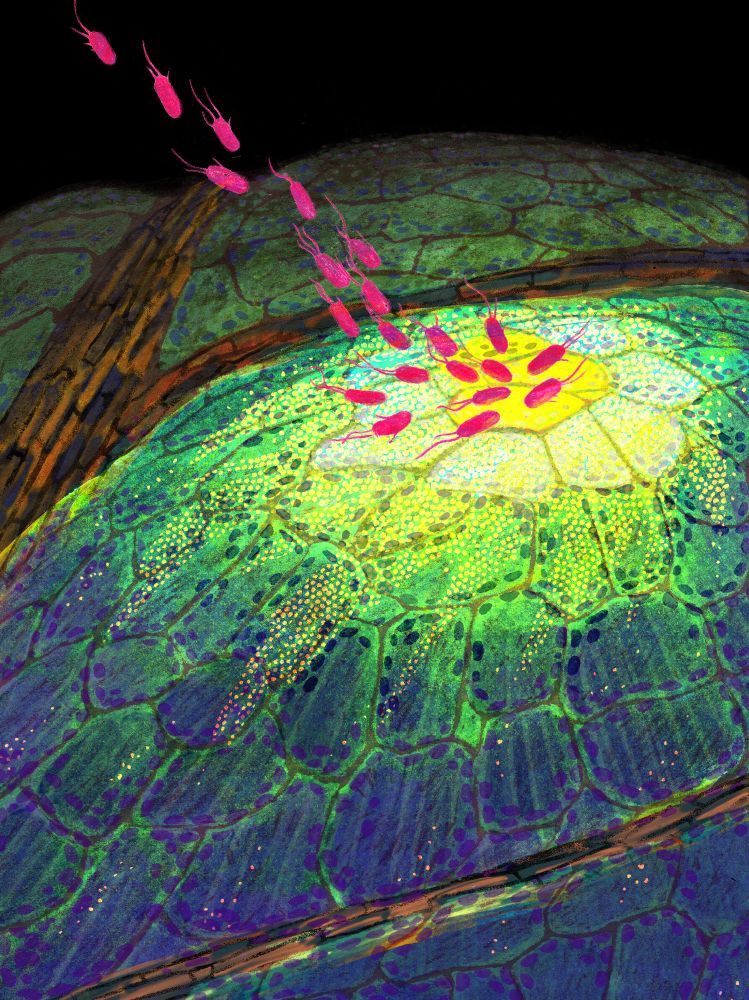
#PlantScience
www.nature.com/articles/s41...
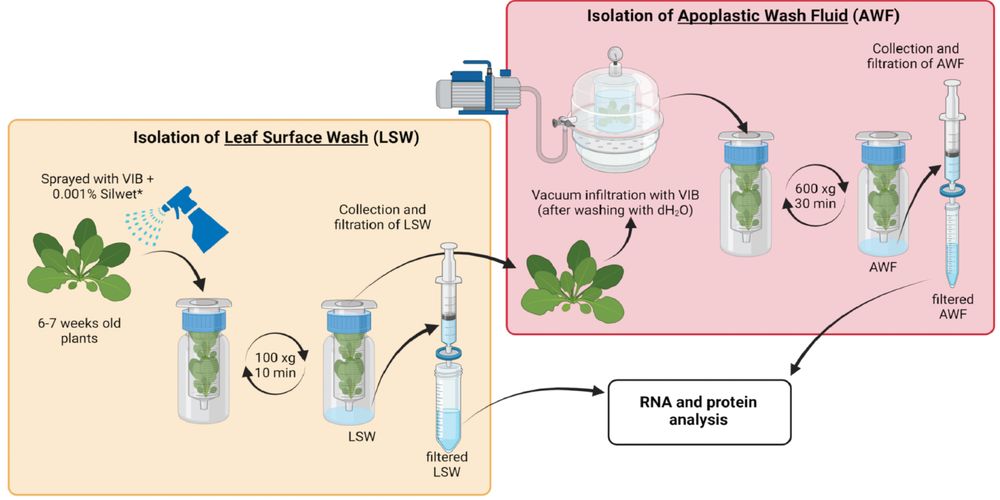
Our latest paper on TE-derived small RNAs is finally out! It reveals a beautiful buffering mechanism involving epigenetics, chromatin organization, and pathogen resistance. A great collaboration with @plantevolution.bsky.social, don’t miss it!
www.nature.com/articles/s41...
Our latest paper on TE-derived small RNAs is finally out! It reveals a beautiful buffering mechanism involving epigenetics, chromatin organization, and pathogen resistance. A great collaboration with @plantevolution.bsky.social, don’t miss it!
www.nature.com/articles/s41...
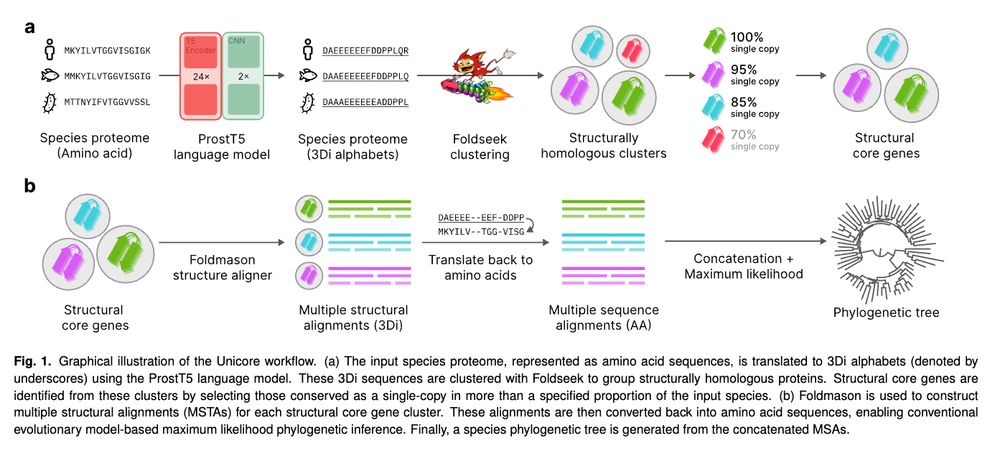
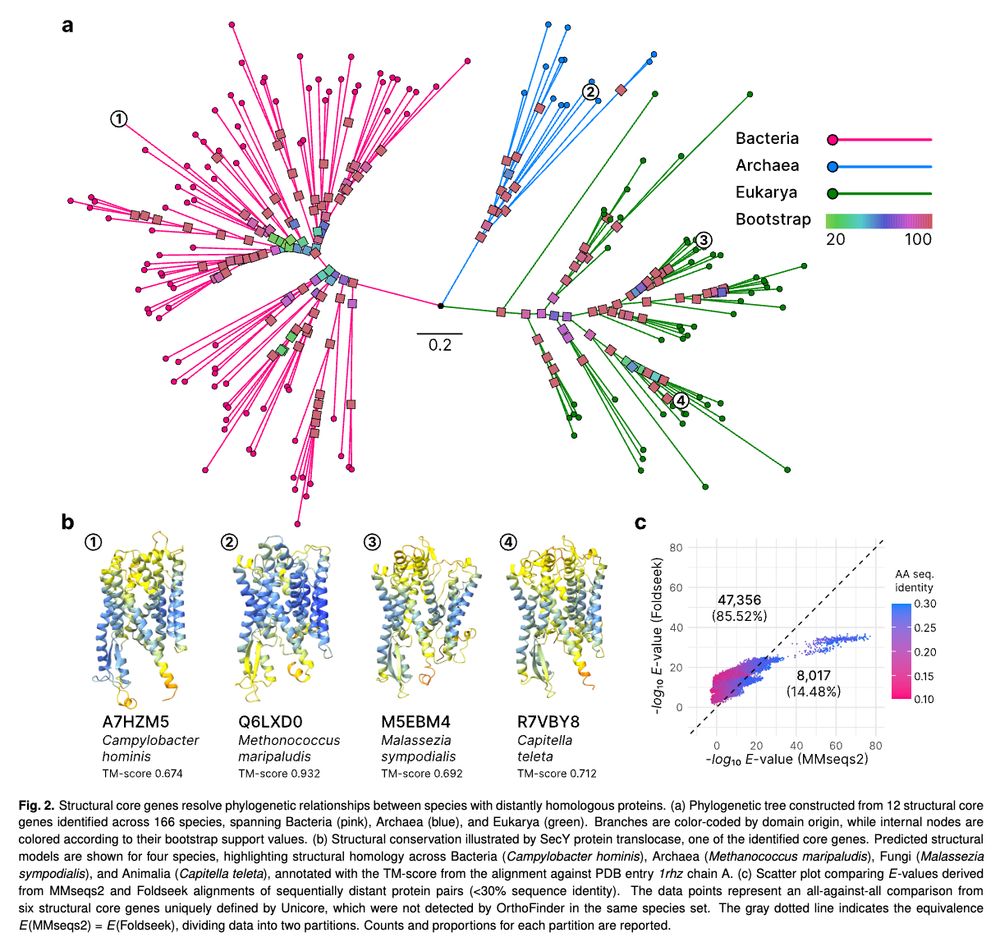
Interested in contributing? Please contact us!
#plantscience
www.biorxiv.org/content/10.1...
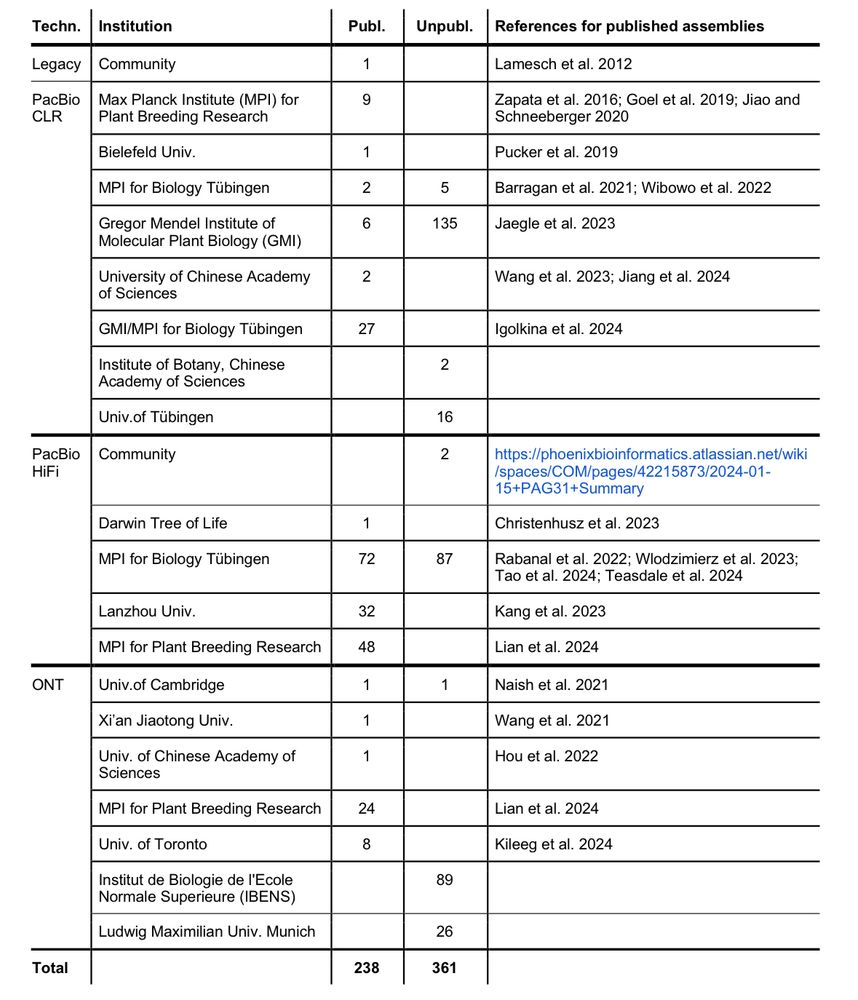
Interested in contributing? Please contact us!
#plantscience
www.biorxiv.org/content/10.1...

🌿 Decipher the molecular mechanisms underlying plant immunity
🌱 Identify & characterise pattern recognition receptors
⏳ Apply soon! Application review already starts 6 Jan '25
www.tsl.ac.uk/working-at-t...

🌿 Decipher the molecular mechanisms underlying plant immunity
🌱 Identify & characterise pattern recognition receptors
⏳ Apply soon! Application review already starts 6 Jan '25
www.tsl.ac.uk/working-at-t...