Mir Lab
@mirlab.bsky.social
650 followers
270 following
29 posts
Lab run by Mustafa Mir studying transcription and nuclear organization using live imaging at University of Pennsylvania, Children's Hospital of Philadelphia, and Howard Hughes Medical Institute. mir-lab.com
Posts
Media
Videos
Starter Packs
Reposted by Mir Lab
Reposted by Mir Lab
Mir Lab
@mirlab.bsky.social
· Apr 11
Mir Lab
@mirlab.bsky.social
· Apr 11
Mir Lab
@mirlab.bsky.social
· Apr 11
Mir Lab
@mirlab.bsky.social
· Apr 9
Mir Lab
@mirlab.bsky.social
· Apr 9
Mir Lab
@mirlab.bsky.social
· Apr 9

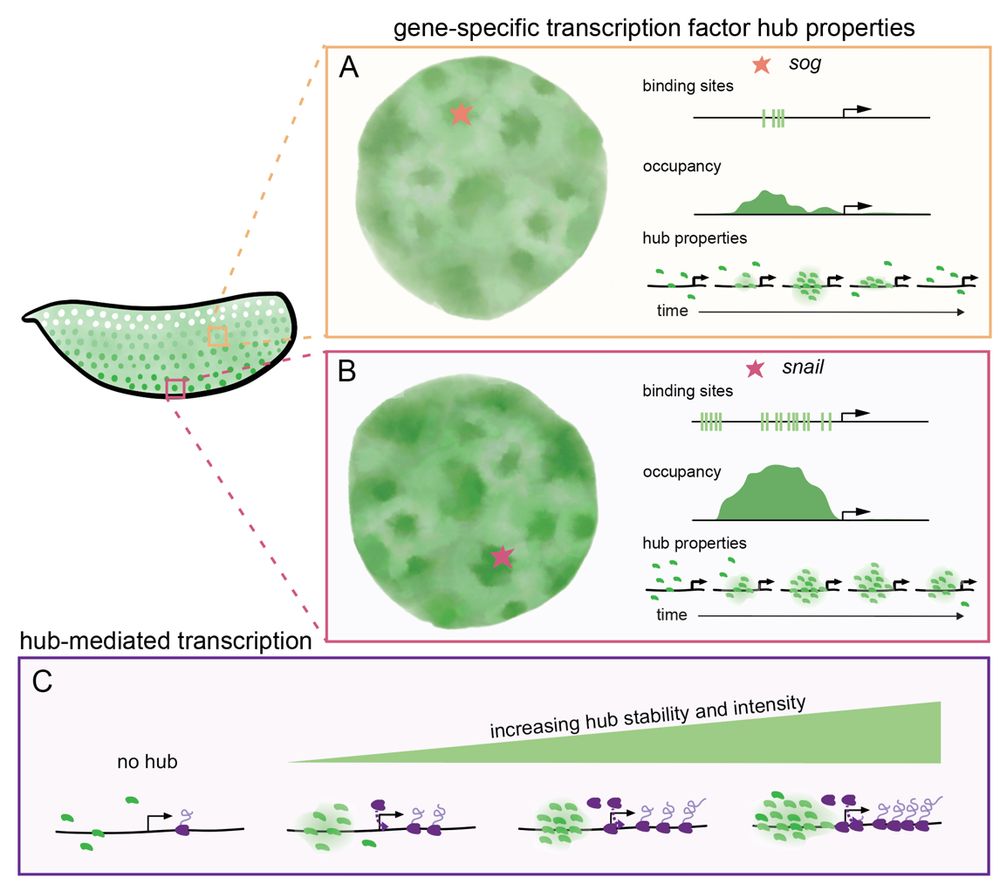
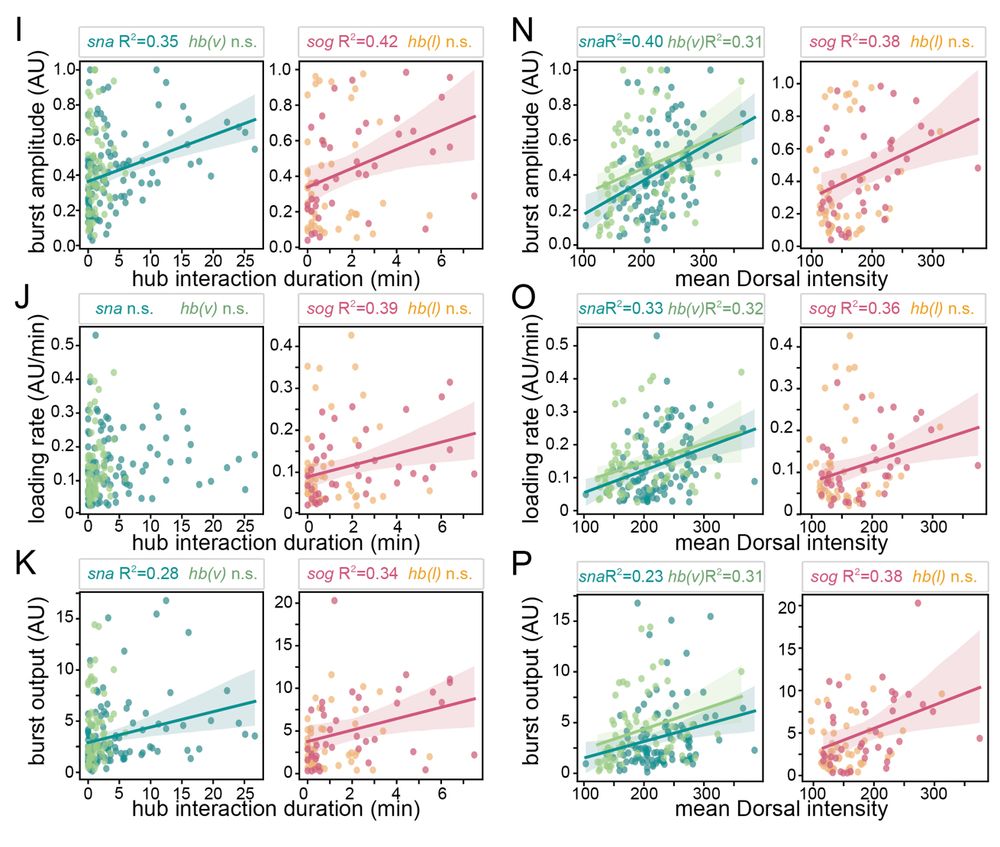
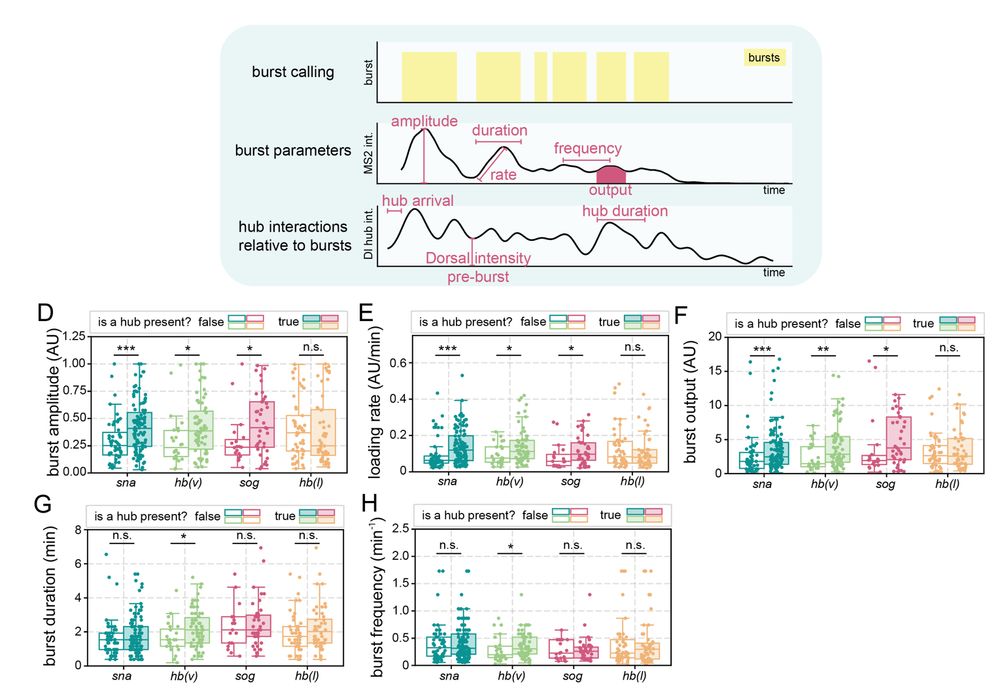
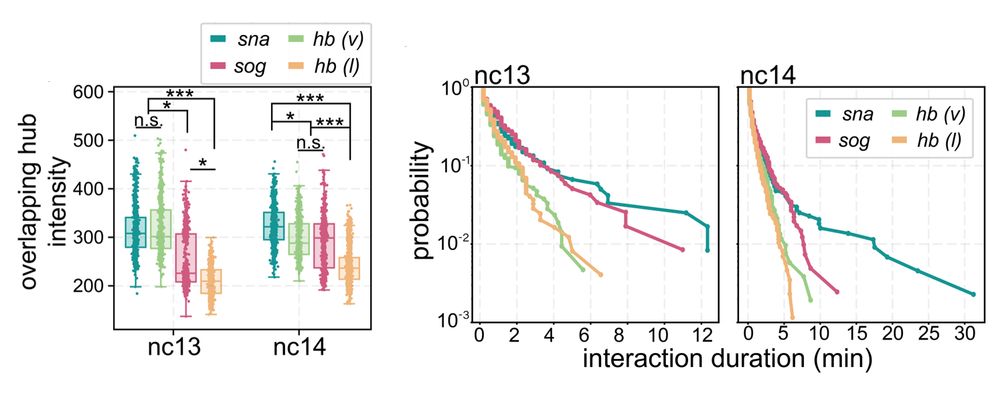
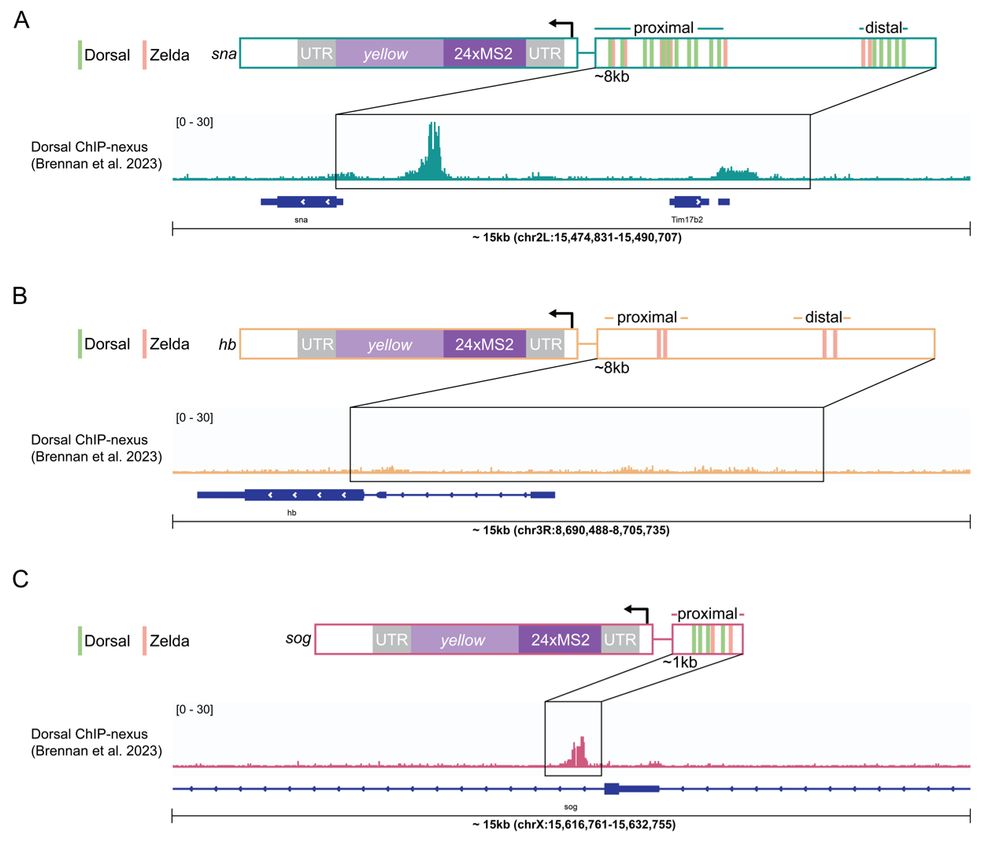
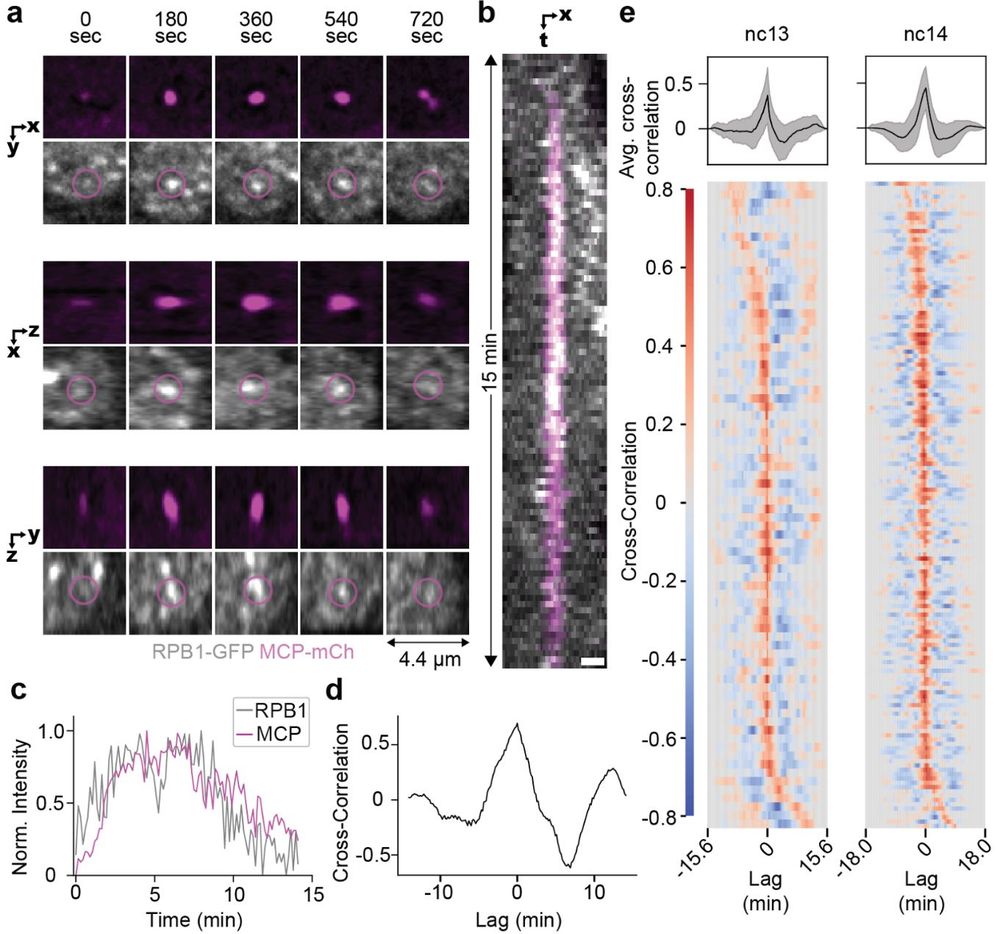
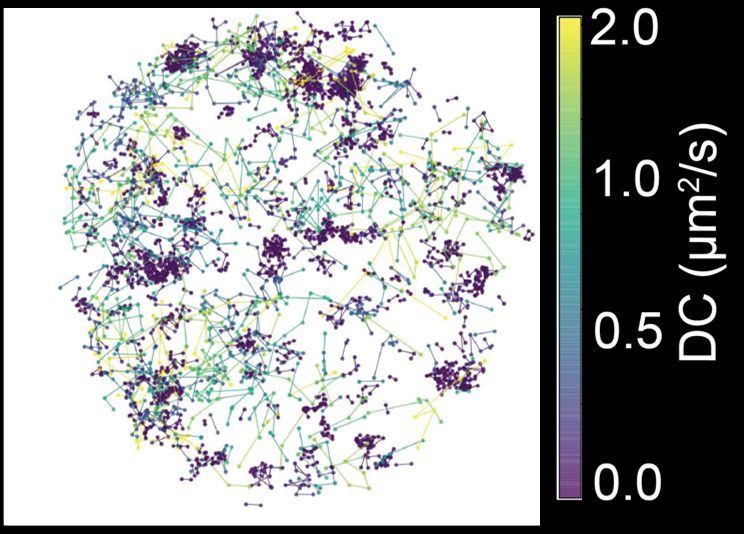
Transcription factor hubs exhibit gene-specific properties that tune expression
The spatial and temporal control of gene expression relies on transcription factors binding to and occupying their target sites. Transcription factor hubs - localized, high-concentration microenvironm...
www.biorxiv.org
Reposted by Mir Lab
Mir Lab
@mirlab.bsky.social
· Feb 12
Mir Lab
@mirlab.bsky.social
· Feb 12
Mir Lab
@mirlab.bsky.social
· Feb 12
Mir Lab
@mirlab.bsky.social
· Feb 12