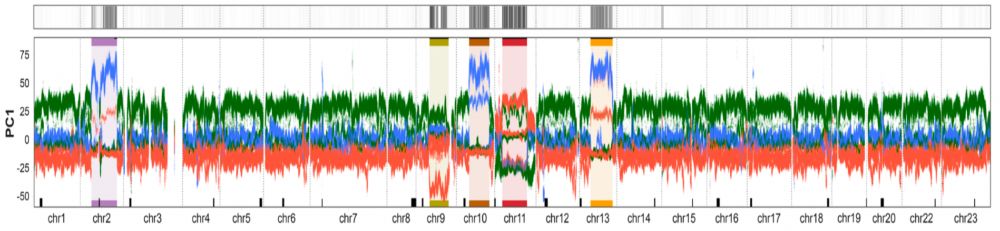
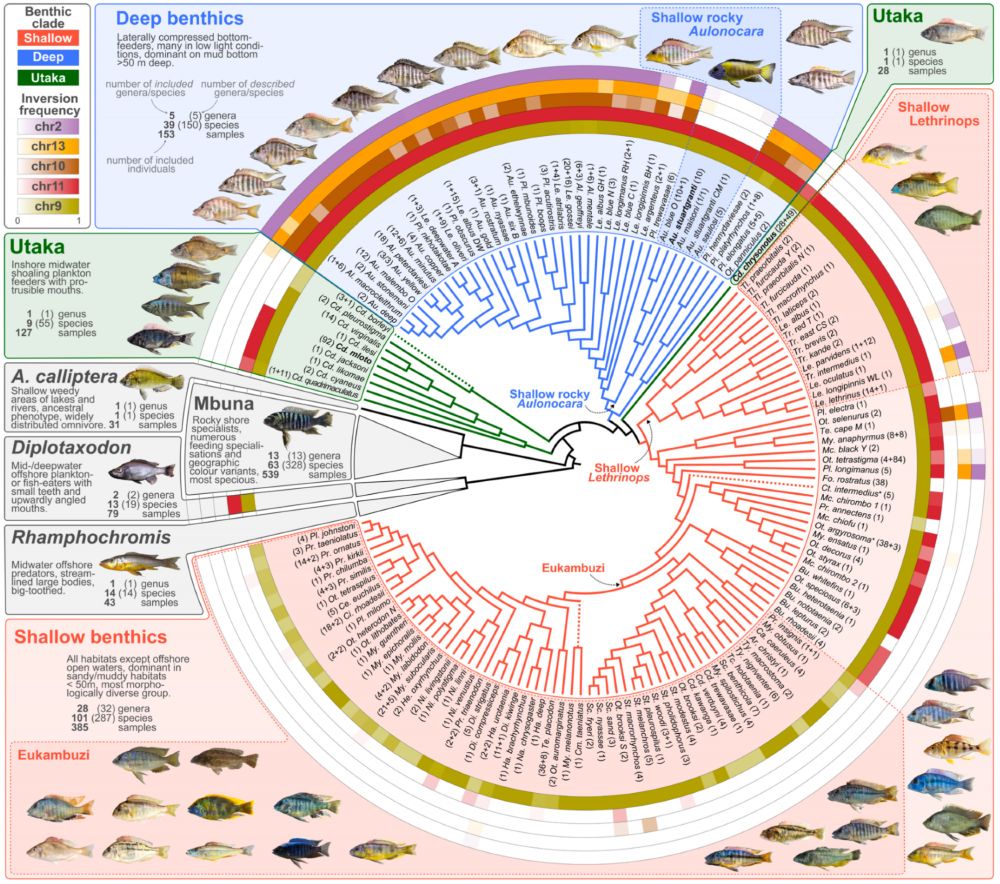
Moritz Blumer
@moritzblumer.bsky.social
140 followers
280 following
16 posts
PhD Student @GeneticsCam. Interested in Genomics, Biodiversity, Speciation, Conservation 🐟🦇🐋🐛🐆. Formerly @goetheuni & @mpicbg.
http://github.com/MoritzBlumer
🇪🇺
Posts
Media
Videos
Starter Packs
Reposted by Moritz Blumer
Moritz Blumer
@moritzblumer.bsky.social
· Jun 19
Moritz Blumer
@moritzblumer.bsky.social
· Jun 19
Moritz Blumer
@moritzblumer.bsky.social
· Jun 19
Moritz Blumer
@moritzblumer.bsky.social
· Jun 19
Moritz Blumer
@moritzblumer.bsky.social
· Jun 19
Moritz Blumer
@moritzblumer.bsky.social
· Jun 19
Moritz Blumer
@moritzblumer.bsky.social
· Jun 19
Moritz Blumer
@moritzblumer.bsky.social
· Jun 19
Moritz Blumer
@moritzblumer.bsky.social
· Jun 19
Reposted by Moritz Blumer
Reposted by Moritz Blumer
Reposted by Moritz Blumer
Reposted by Moritz Blumer
Milan Malinsky
@millanek.bsky.social
· Mar 12
Reposted by Moritz Blumer
Anne Applebaum
@anneapplebaum.bsky.social
· Feb 22