Narayanan Lab
@narayananlab.bsky.social
180 followers
35 following
48 posts
Systems neuroscientist & Parkinson's neurologist. We map brain circuits of higher-order thought.
@UIowaNeuro
@IowaNeurology
Posts
Media
Videos
Starter Packs
Narayanan Lab
@narayananlab.bsky.social
· May 19
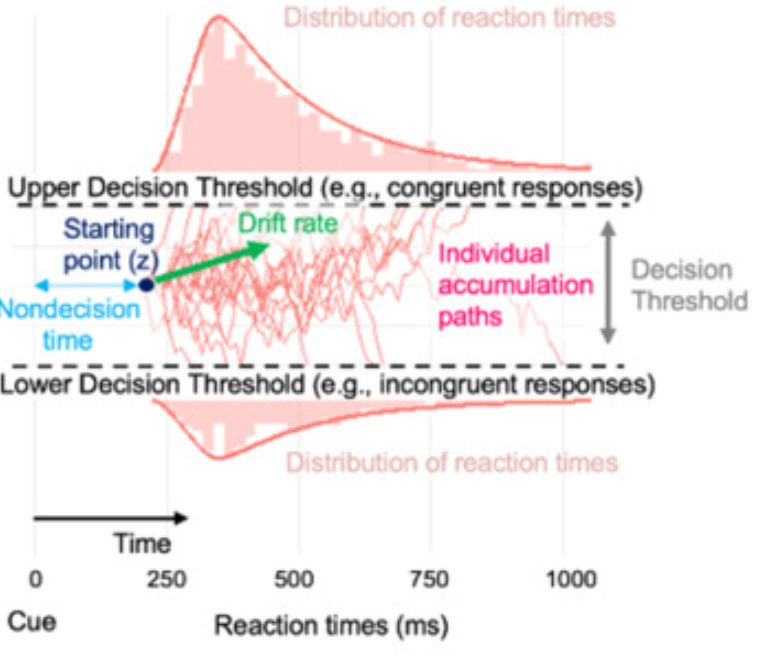
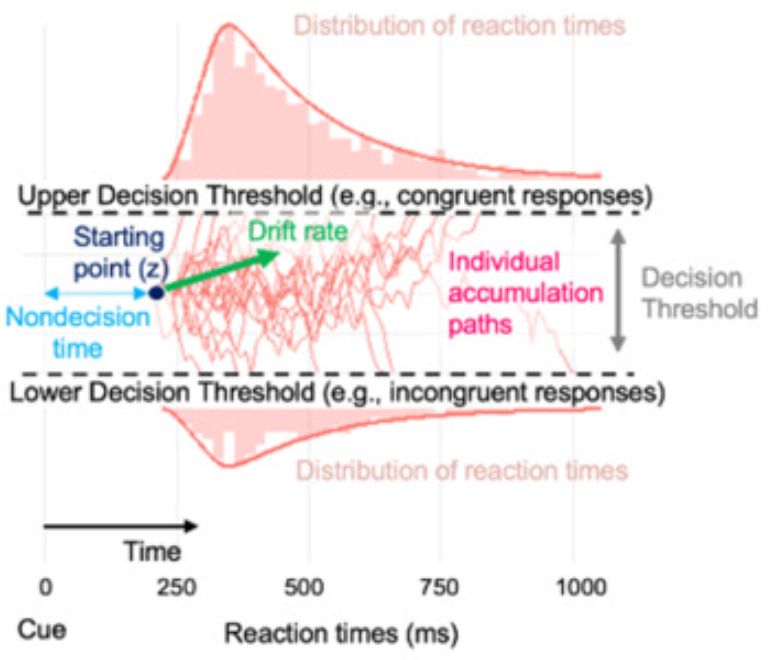
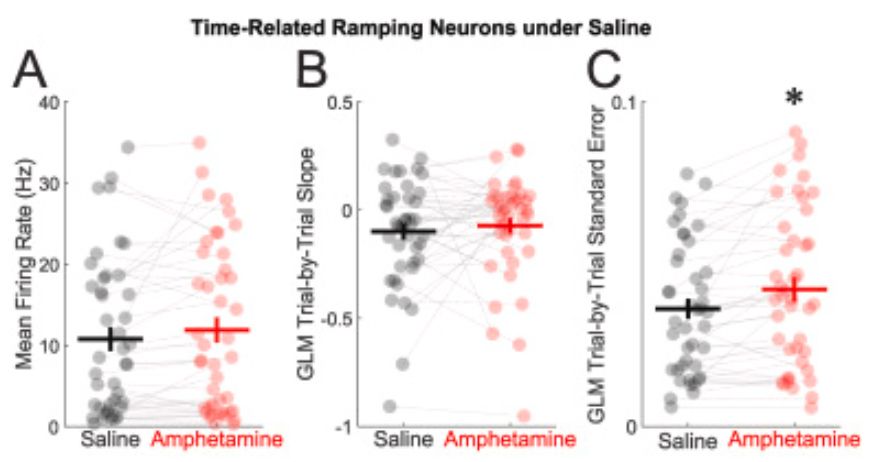
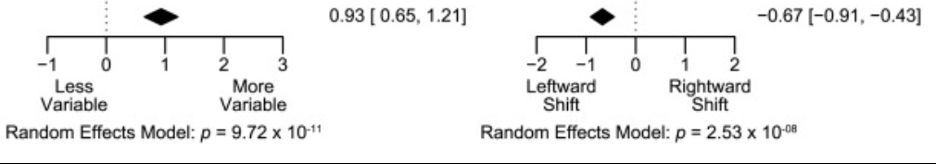
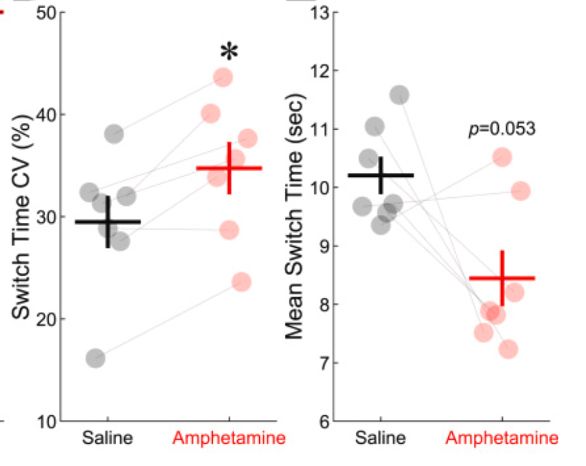
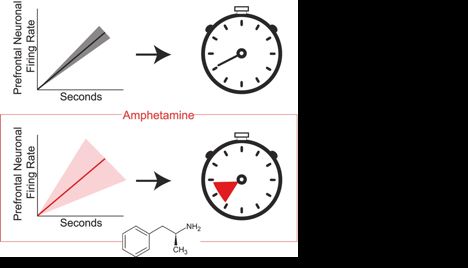
Amphetamine scrambles the brain's sense of time by degrading prefrontal neuron coordination
Researchers have found that amphetamine alters how the brain processes time, increasing variability in the activity of neurons that encode temporal information. The study provides insight into how the...
www.psypost.org
Narayanan Lab
@narayananlab.bsky.social
· May 14

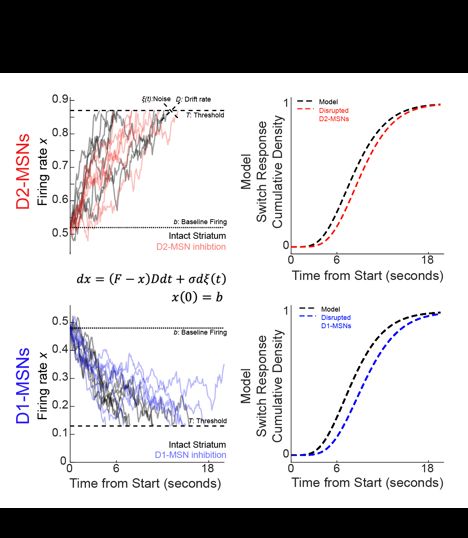
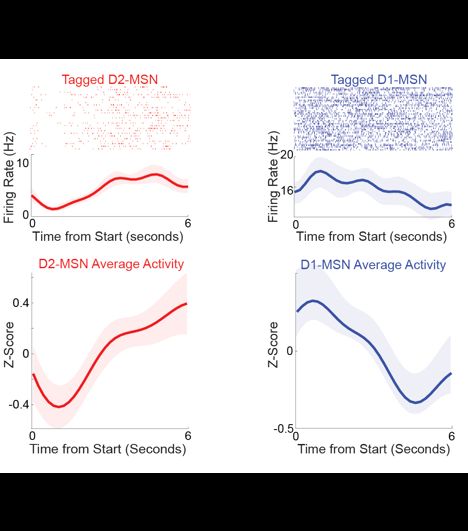
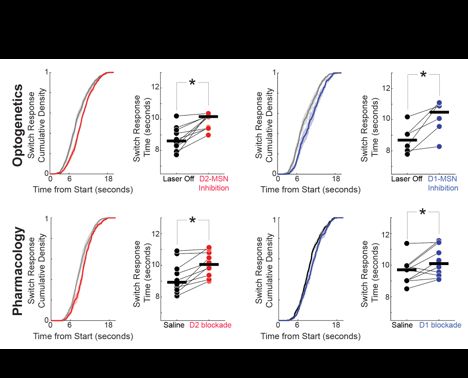
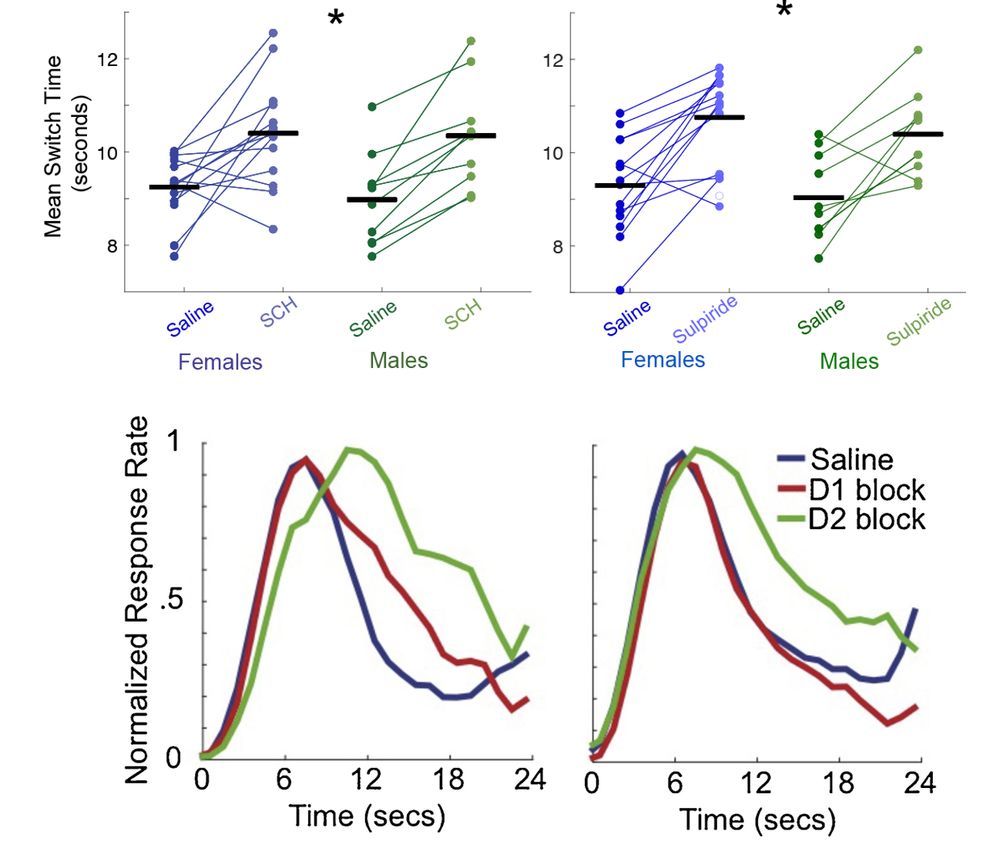
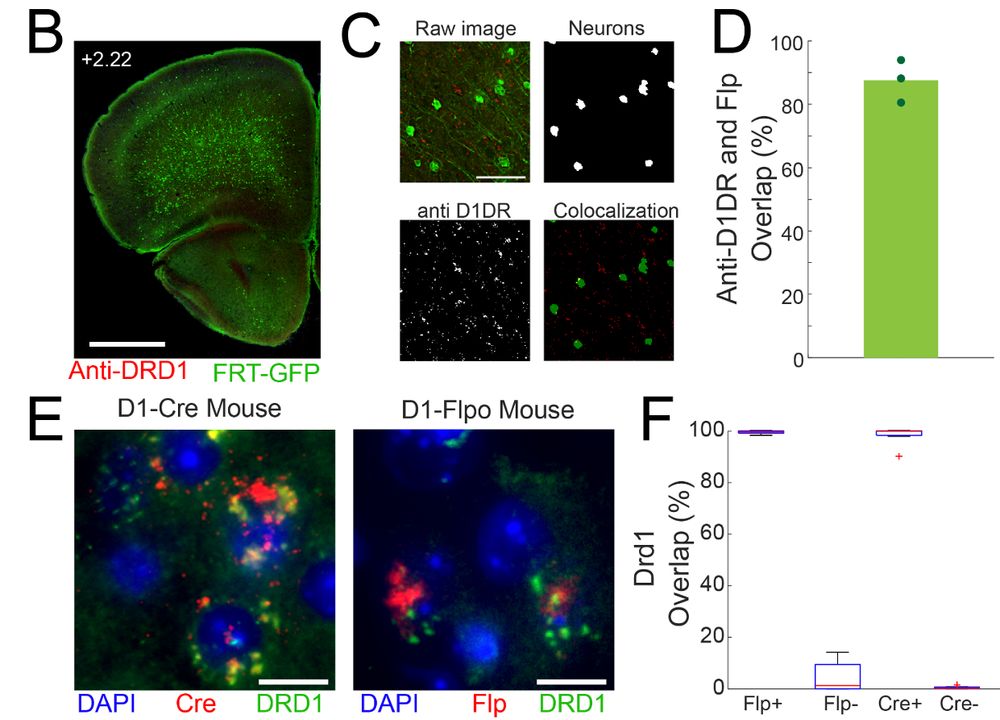
Diverse activity in prefrontal projections promotes temporal control of action
Prefrontal neurons can have diverse activity during cognitive functions like working memory, attention, and timing; however, the importance of this heterogeneity is unclear. Our goal was to better und...
www.biorxiv.org