Jonas Neipel
@neipel.bsky.social
90 followers
160 following
11 posts
Theoretical biophysicists in Grill and Jülicher labs. (MPI-CBG, MPI-PKS Dresden) Interested in anything from genome regulation to developmental biology, currently focusing on left-right symmetry breaking and shape/curvature/ geometry sensing.
Posts
Media
Videos
Starter Packs
Reposted by Jonas Neipel
Jonas Neipel
@neipel.bsky.social
· Jul 20
Jonas Neipel
@neipel.bsky.social
· Jul 20
Jonas Neipel
@neipel.bsky.social
· Jul 20

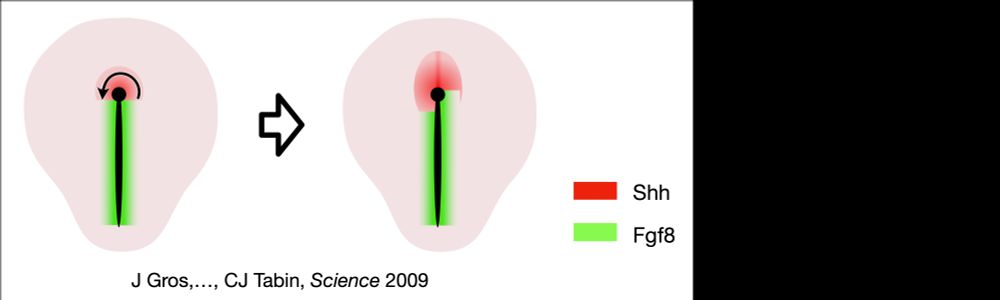
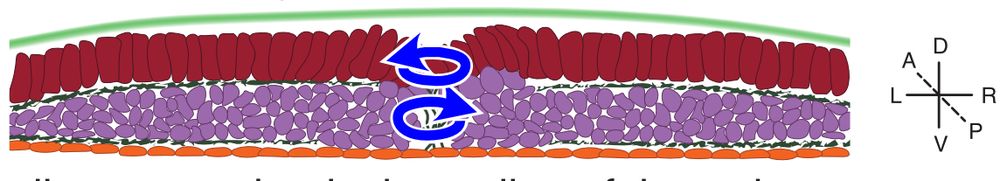
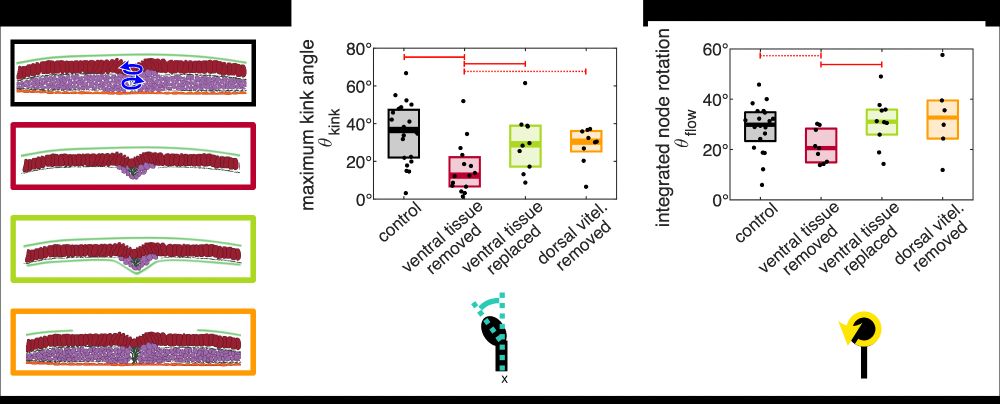
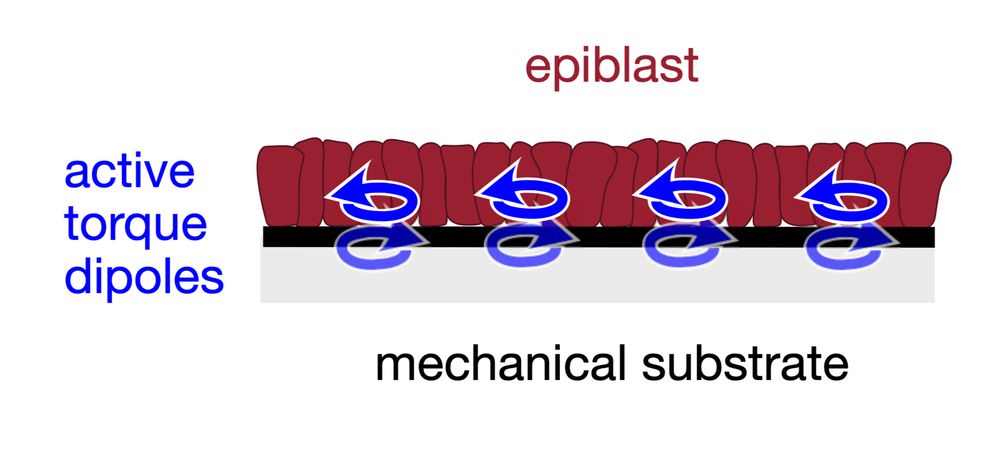
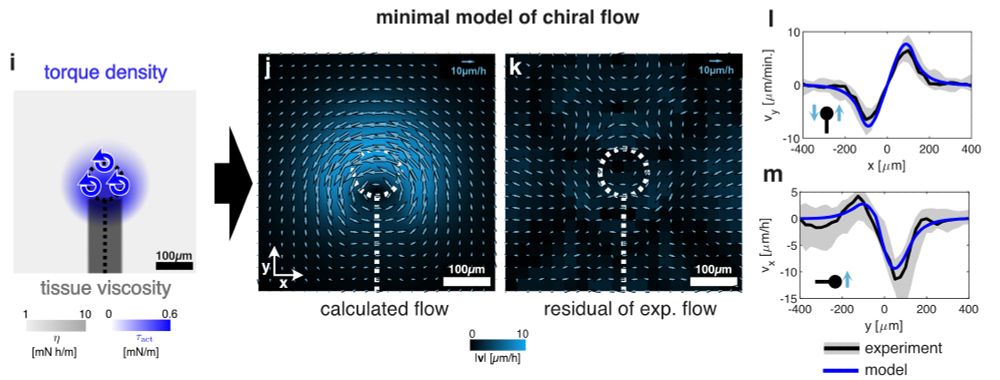
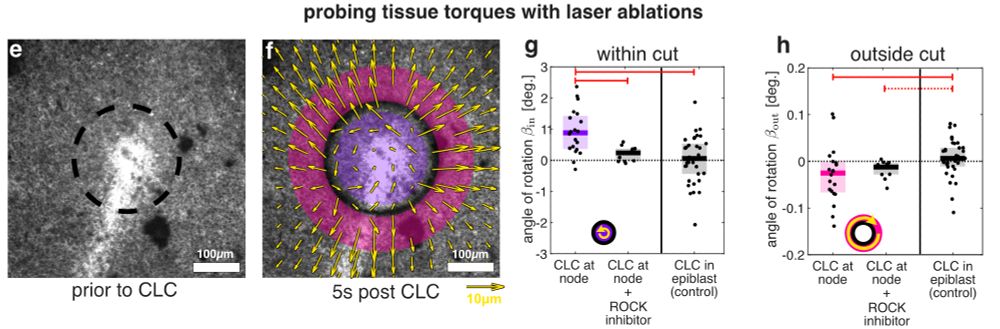
An active torque dipole across tissue layers drives avian left-right symmetry breaking
Unlike in mice, frogs, and fish, left-right (L/R) body axis formation in avian embryos does not arise from the chiral beat of cilia. Instead, a counter-clockwise tissue rotation around Hensen′ node, t...
doi.org