Noah Fierer
@noahfierer.bsky.social
2.6K followers
200 following
84 posts
Microbial ecologist
https://www.fiererlab.org/
Posts
Media
Videos
Starter Packs
Noah Fierer
@noahfierer.bsky.social
· Aug 6

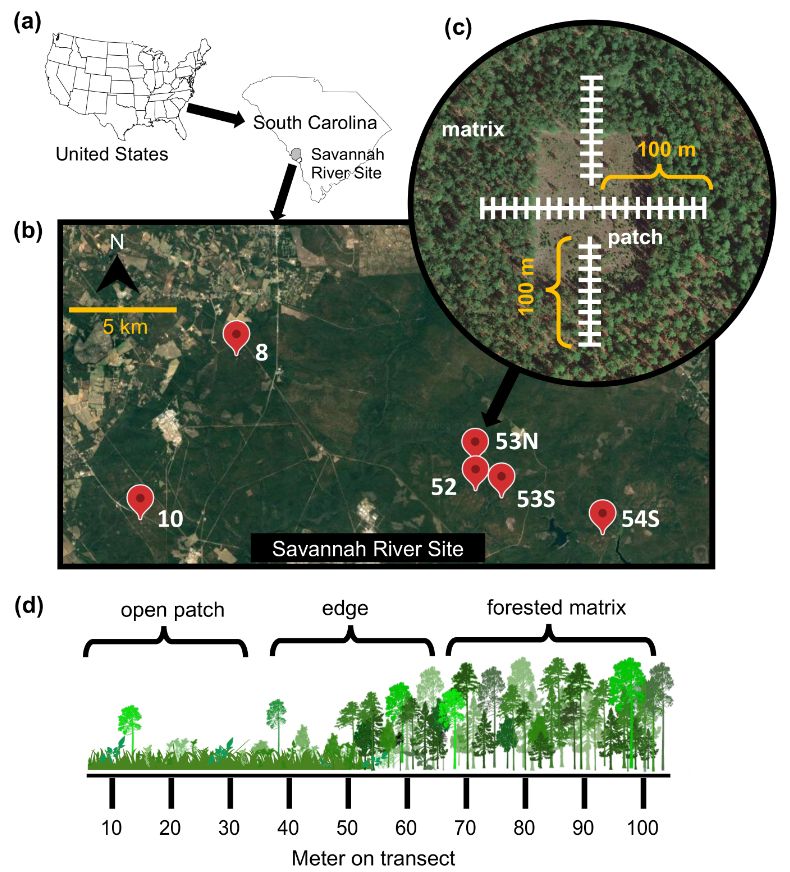
A cross-environment comparison of nontuberculous mycobacterial diversity
Nontuberculous mycobacteria (NTM) are a group of environmental bacteria that encompass nearly 200 described species, some of which can cause chronic pulmonary infection in humans. What makes these inf...
www.biorxiv.org
Noah Fierer
@noahfierer.bsky.social
· Jul 18
Noah Fierer
@noahfierer.bsky.social
· Jul 18
Noah Fierer
@noahfierer.bsky.social
· Jul 18
Noah Fierer
@noahfierer.bsky.social
· Jul 18
Noah Fierer
@noahfierer.bsky.social
· Jul 18
Noah Fierer
@noahfierer.bsky.social
· Jul 18

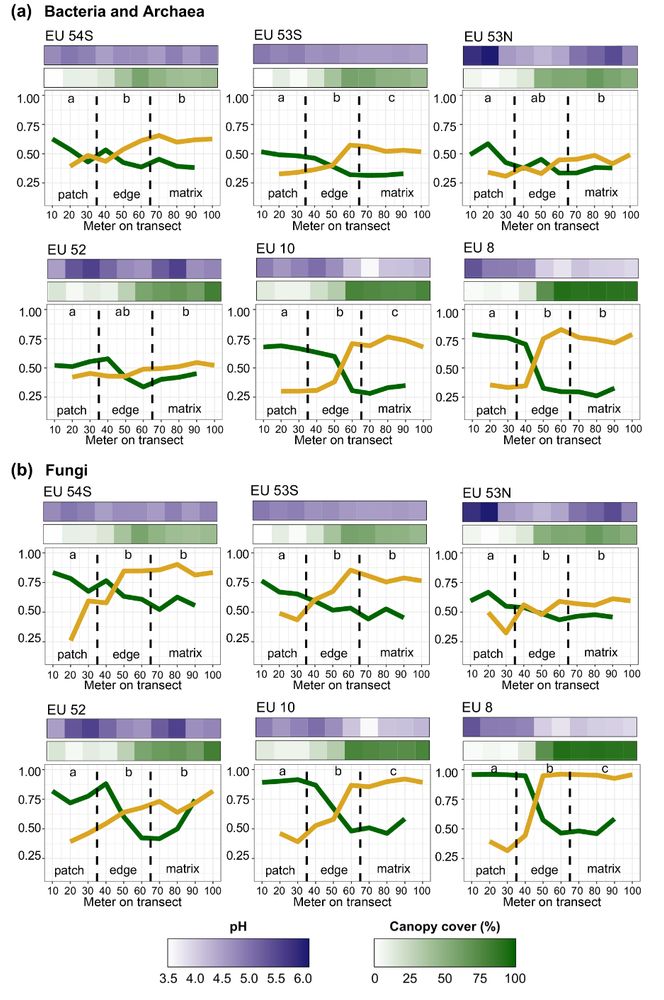
A periodic table of bacteria?: Mapping bacterial diversity in trait space
Bacterial diversity can be overwhelming. There is an ever-expanding number of bacterial taxa being discovered, but many of these taxa remain uncharacterized with unknown traits and environmental prefe...
www.biorxiv.org
Noah Fierer
@noahfierer.bsky.social
· Jul 14
Noah Fierer
@noahfierer.bsky.social
· Jun 25
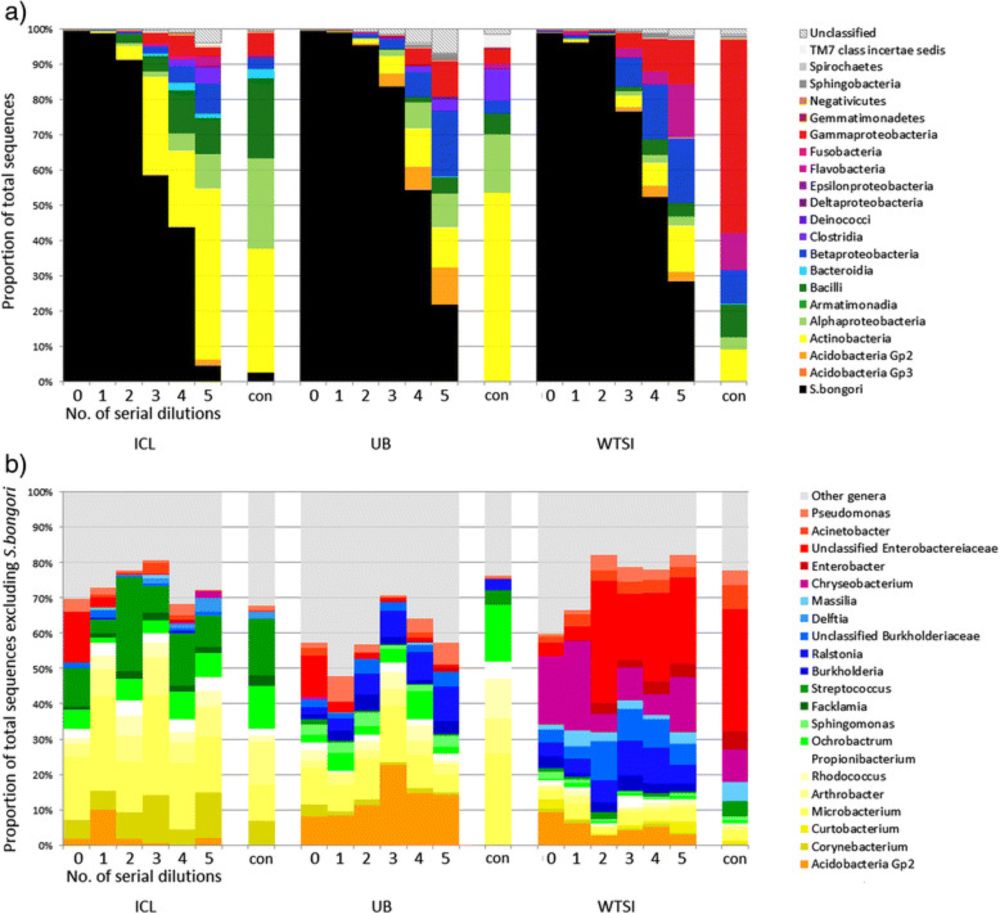
Reagent and laboratory contamination can critically impact sequence-based microbiome analyses - BMC Biology
Background The study of microbial communities has been revolutionised in recent years by the widespread adoption of culture independent analytical techniques such as 16S rRNA gene sequencing and metag...
bmcbiol.biomedcentral.com
Noah Fierer
@noahfierer.bsky.social
· Jun 24
Noah Fierer
@noahfierer.bsky.social
· Jun 21
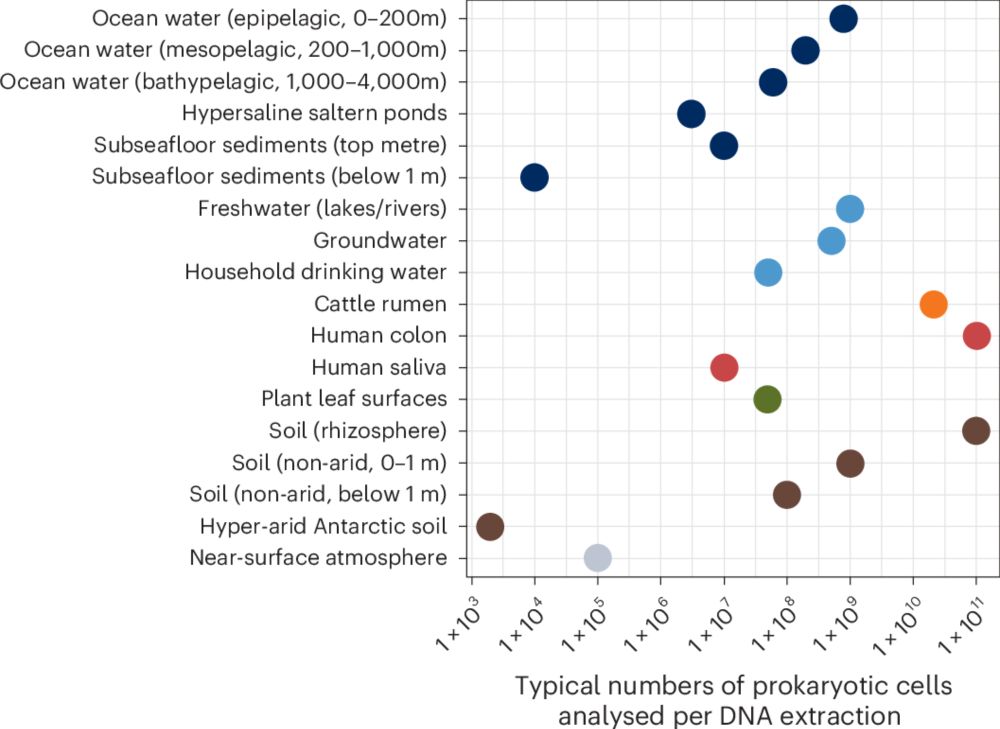
Guidelines for preventing and reporting contamination in low-biomass microbiome studies
Nature Microbiology - In this Consensus Statement, the authors outline strategies for processing, analysing and interpreting low-biomass microbiome samples, and provide recommendations to minimize...
rdcu.be
Noah Fierer
@noahfierer.bsky.social
· May 30

Continent-wide assessment of the strain-level diversity of Bradyrhizobium, a dominant soil bacterial genus
Global surveys of soil bacteria have identified several taxa that are nearly ubiquitous and often the most abundant members of soil bacterial communities. However, it remains unclear why these taxa ar...
www.biorxiv.org
Noah Fierer
@noahfierer.bsky.social
· May 21

Bacteria break through one-micrometer-square passages by flagellar wrapping
Confined spaces are omnipresent in the micro-environments, including soil aggregates and intestinal crypts, yet little is known about how bacteria behave under such conditions where movement is challe...
www.biorxiv.org
Noah Fierer
@noahfierer.bsky.social
· May 8
Noah Fierer
@noahfierer.bsky.social
· May 7
Noah Fierer
@noahfierer.bsky.social
· May 7
Noah Fierer
@noahfierer.bsky.social
· May 1
Noah Fierer
@noahfierer.bsky.social
· Apr 29
Reposted by Noah Fierer
Noah Fierer
@noahfierer.bsky.social
· Feb 28
Noah Fierer
@noahfierer.bsky.social
· Feb 22