Noémie Lefrancq
@noemielefrancq.bsky.social
730 followers
250 following
20 posts
Phylogenetics 🧬 , infectious disease dynamics 🦠 & modelling 💻 #IDSky #IDModelling
Postdoctoral fellow @ETH Zürich (in Basel)
Previously @Cambridge_Uni and @institutpasteur
Posts
Media
Videos
Starter Packs
Reposted by Noémie Lefrancq
Reposted by Noémie Lefrancq
Moritz Kraemer
@mugkraemer.bsky.social
· Feb 20
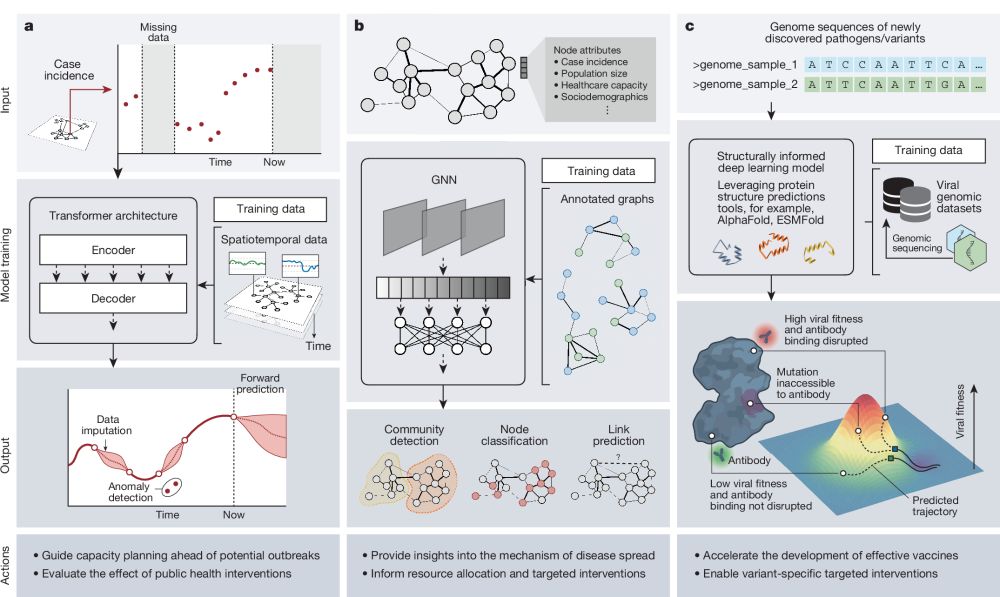
Artificial intelligence for modelling infectious disease epidemics
Nature - This Perspective considers the application to infectious disease modelling of AI systems that combine machine learning, computational statistics, information retrieval and data science.
rdcu.be
Reposted by Noémie Lefrancq
Richard Neher
@neher.io
· Jan 11

The mutation rate of SARS-CoV-2 is highly variable between sites and is influenced by sequence context, genomic region, and RNA structure
RNA viruses like SARS-CoV-2 have a high mutation rate, which contributes to their rapid evolution. The rate of mutations depends on the mutation type (e.g., A→C, A→G, etc.) and can vary between sites ...
www.biorxiv.org
Reposted by Noémie Lefrancq
Zannah Salter
@zannahdu.bsky.social
· Dec 2

The brain microbiome: could understanding it help prevent dementia?
Long thought to be sterile, our brains are now believed to harbour all sorts of micro-organisms, from bacteria to fungi. How big a part do they play in Alzheimer’s and similar diseases?
www.theguardian.com
Reposted by Noémie Lefrancq