Nogales Lab
@nogaleslab.bsky.social
1.7K followers
39 following
34 posts
The Nogales Lab @ UC Berkeley. #CryoEM #Transcription #Microtubules #ChromatinModification Bluesky account managed by lab members.
Posts
Media
Videos
Starter Packs
Nogales Lab
@nogaleslab.bsky.social
· Jul 7
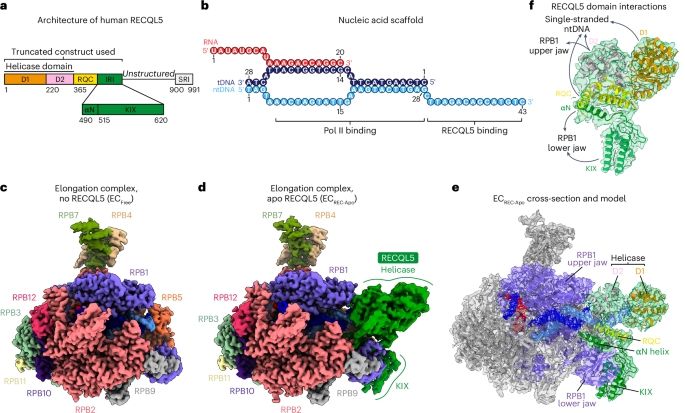
Structural insights into transcriptional regulation by the helicase RECQL5 - Nature Structural & Molecular Biology
Florez Ariza and Lue et al. use cryo-electron microscopy to investigate how the RECQL5 helicase regulates transcription. Their structural findings suggest that RECQL5 can modulate RNA polymerase II’s ...
www.nature.com
Nogales Lab
@nogaleslab.bsky.social
· Jan 31
Nogales Lab
@nogaleslab.bsky.social
· Jan 31
Nogales Lab
@nogaleslab.bsky.social
· Jan 31

Structural insights into transcriptional regulation by the helicase RECQL5
Transcription and its regulation pose a major challenge for genome stability. The helicase RECQL5 has been proposed as an important factor to help safeguard the genome, and is the only member of the h...
www.biorxiv.org


























