Nora Pyenson
@norapyenson.bsky.social
480 followers
97 following
11 posts
Postdoc at NYU
www.norapyenson.com
Posts
Media
Videos
Starter Packs
Reposted by Nora Pyenson
Simon Roux
@simrouxvirus.bsky.social
· Jul 22
Nora Pyenson
@norapyenson.bsky.social
· Dec 12
Nora Pyenson
@norapyenson.bsky.social
· Dec 12
Nora Pyenson
@norapyenson.bsky.social
· Dec 12
Nora Pyenson
@norapyenson.bsky.social
· Dec 12
Nora Pyenson
@norapyenson.bsky.social
· Dec 12
Nora Pyenson
@norapyenson.bsky.social
· Dec 12
Nora Pyenson
@norapyenson.bsky.social
· Dec 12
Nora Pyenson
@norapyenson.bsky.social
· Dec 12
Nora Pyenson
@norapyenson.bsky.social
· Dec 12
Nora Pyenson
@norapyenson.bsky.social
· Dec 12
Nora Pyenson
@norapyenson.bsky.social
· Dec 12
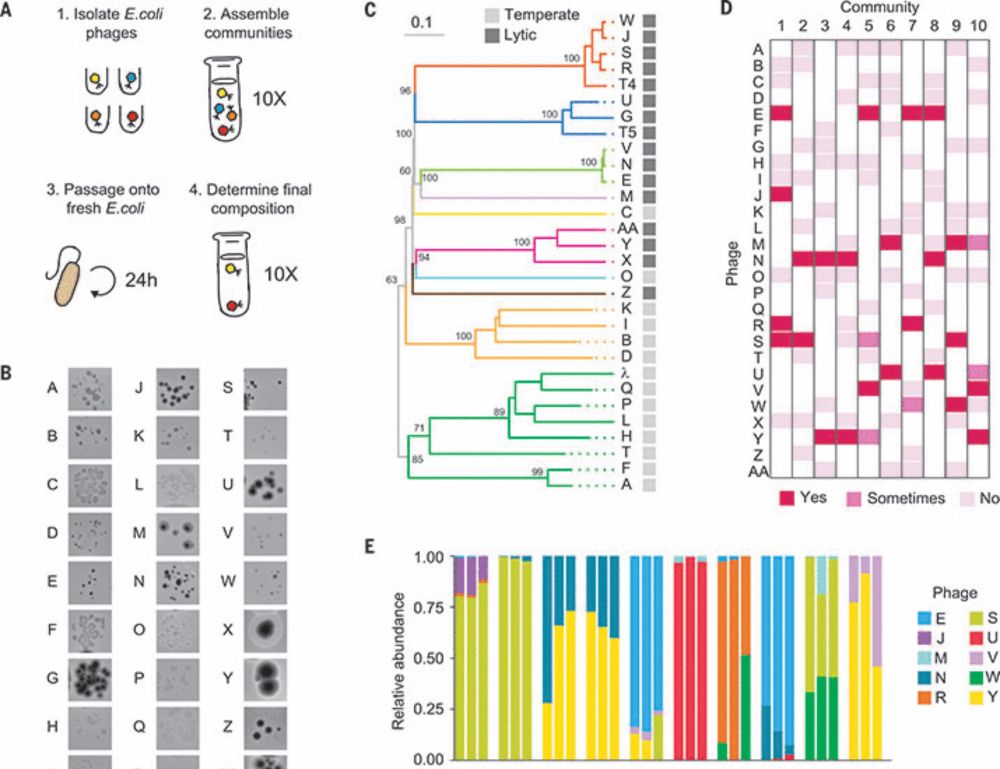
Diverse phage communities are maintained stably on a clonal bacterial host
Bacteriophages are the most abundant and phylogenetically diverse biological entities on Earth, yet the ecological mechanisms that sustain this extraordinary diversity remain unclear. In this study, w...
www.science.org
Reposted by Nora Pyenson
Reposted by Nora Pyenson
Reposted by Nora Pyenson
Noah Houpt
@noahhoupt.bsky.social
· Oct 16


