Posts
Media
Videos
Starter Packs
Pinned
Reposted by Open Free Energy
Open Free Energy
@openfree.energy
· Aug 27
Open Free Energy
@openfree.energy
· Aug 27
Open Free Energy
@openfree.energy
· Aug 19
Open Free Energy
@openfree.energy
· Jun 24
Reposted by Open Free Energy
David L. Dotson
@dotsdl.bsky.social
· May 27
Open Free Energy
@openfree.energy
· May 15
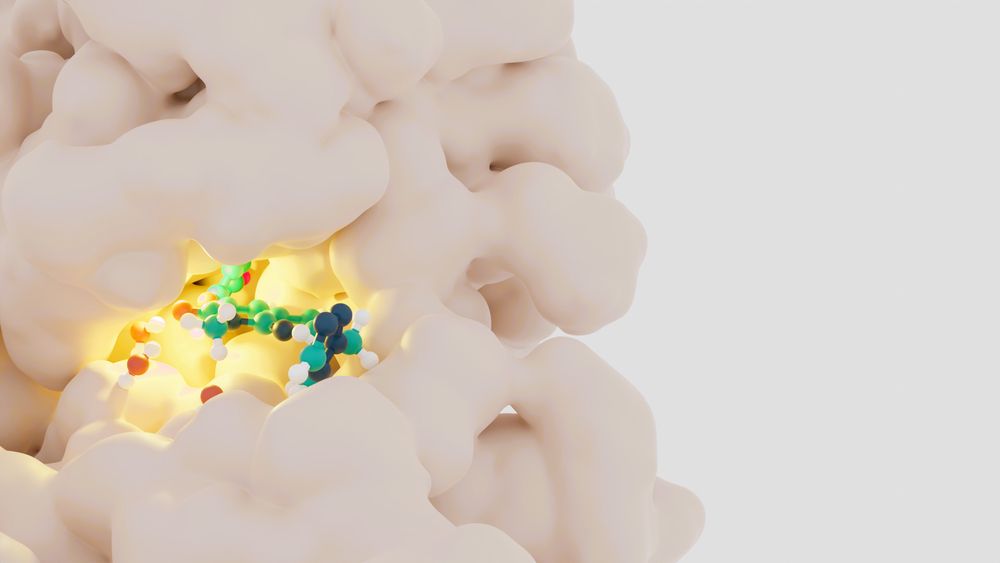
The Free Energy of Everything: Benchmarking OpenFE
Written by: Josh Horton, PhD
Who is OpenFE and what do they do?
Open Free Energy (OpenFE) sits at the nexus of academia and industry. We are developing an open-source software ecosystem for alchem...
blog.omsf.io
Reposted by Open Free Energy
Reposted by Open Free Energy
Open Free Energy
@openfree.energy
· Apr 30
Open Free Energy
@openfree.energy
· Apr 1
Open Free Energy
@openfree.energy
· Apr 1
Open Free Energy
@openfree.energy
· Apr 1

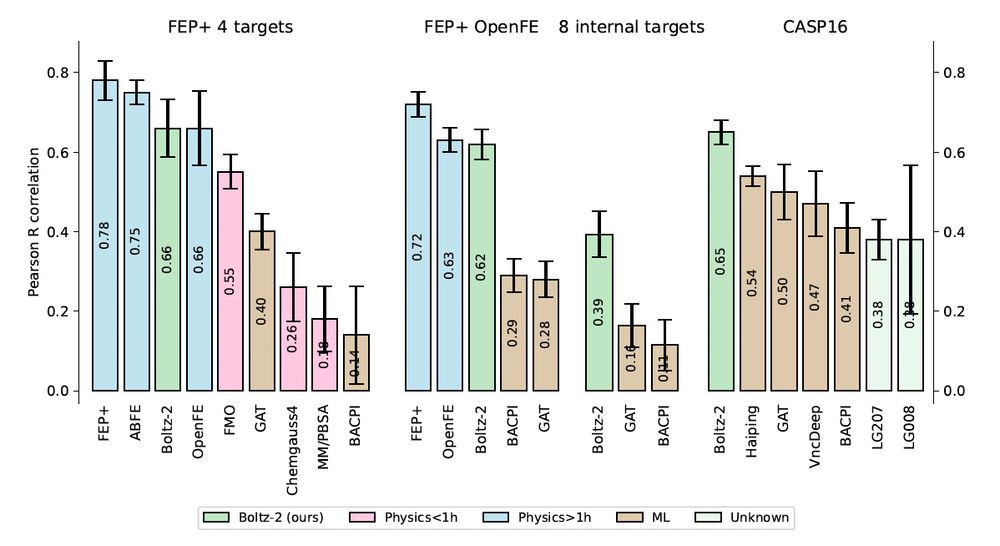
Leveraging Alchemical Free Energy Calculations with Accurate Protein Structure Prediction
Small-molecule lead optimisation in early-stage drug discovery is broadly supported by computational chemistry approaches throughout industry. Over the last decade, Free Energy Perturbation (FEP) has ...
chemrxiv.org
Open Free Energy
@openfree.energy
· Mar 4