@phil-fradkin.bsky.social
520 followers
340 following
10 posts
Born too late to explore the Earth
Too early to explore the Galaxy
Just in time to model the cell and climb some rocks
PhD @ UofT and Vector Institute
Posts
Media
Videos
Starter Packs
Reposted
Reposted
Reposted
Quaid Morris
@quaidmorris.bsky.social
· Jun 3

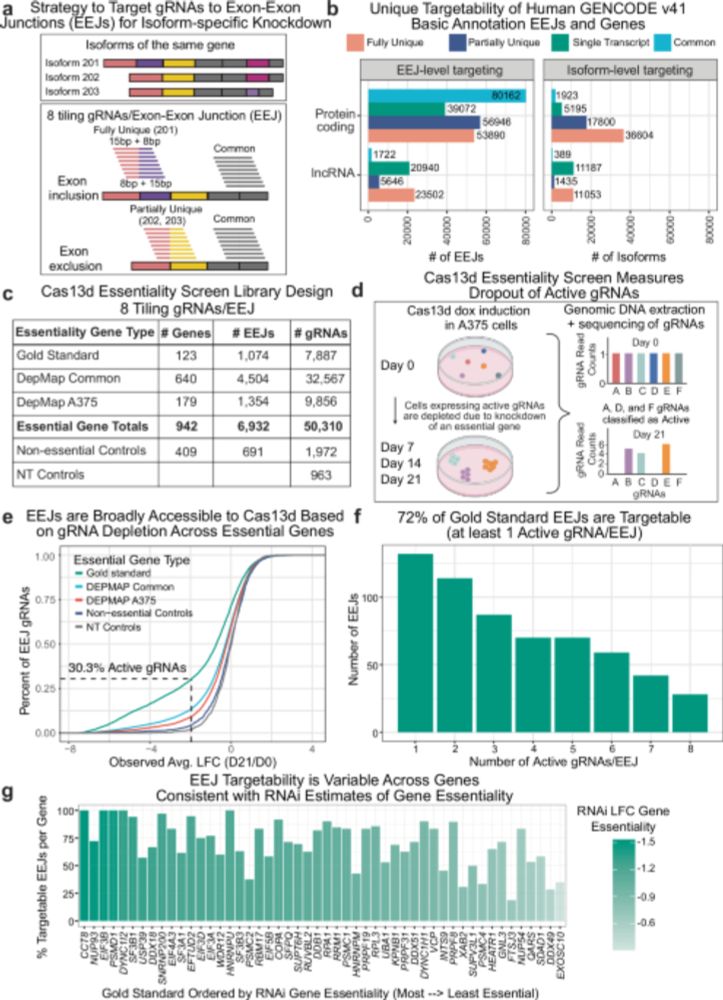
Damage and Misrepair Signatures: Compact Representations of Pan-cancer Mutational Processes
Mutational signatures of single-base substitutions (SBSs) characterize somatic mutation processes which contribute to cancer development and progression. However, current mutational signatures do not ...
www.biorxiv.org
Reposted
Reposted
Reposted
Reposted