Peter Koo
@pkoo562.bsky.social
3.1K followers
1.3K following
110 posts
AI4Science researcher. Associate Professor @CSHL. My lab advances AI for genomics and healthcare!
http://koo-lab.github.io
Posts
Media
Videos
Starter Packs
Peter Koo
@pkoo562.bsky.social
· Jul 16
Peter Koo
@pkoo562.bsky.social
· Jul 16
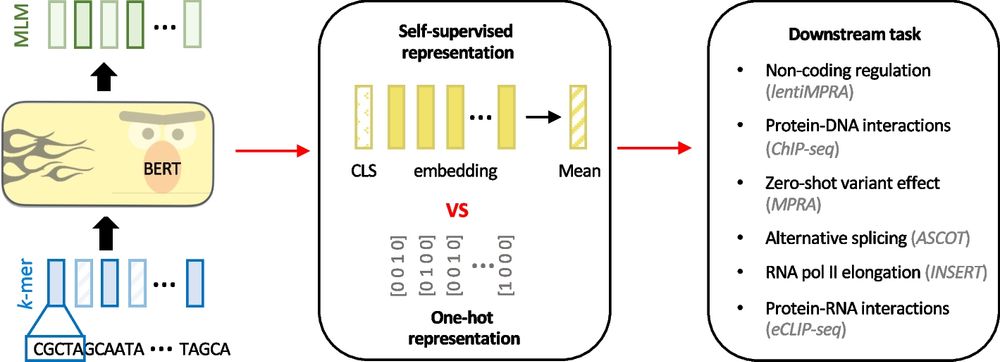
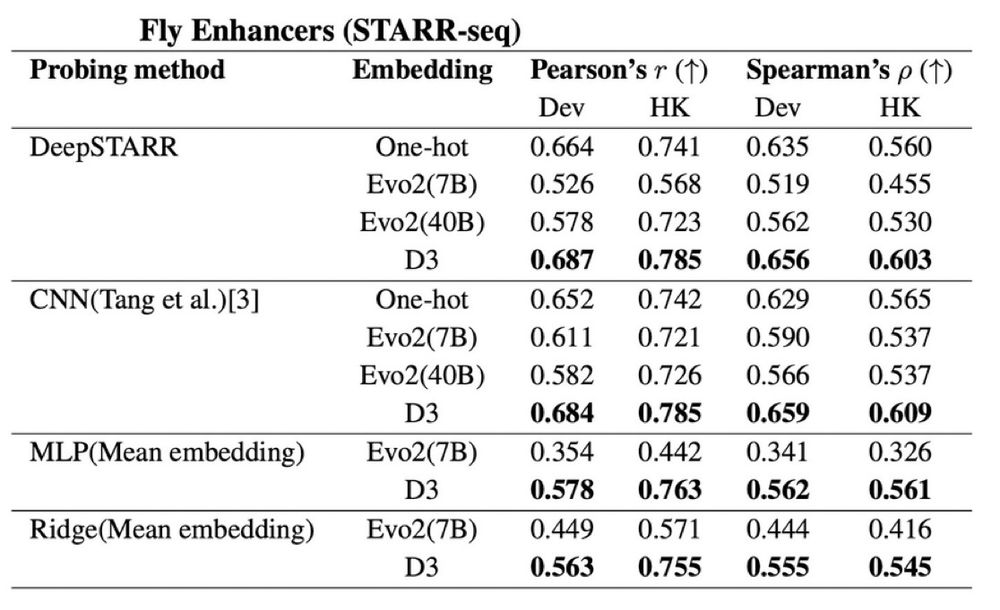
Evaluating the representational power of pre-trained DNA language models for regulatory genomics - Genome Biology
Background The emergence of genomic language models (gLMs) offers an unsupervised approach to learning a wide diversity of cis-regulatory patterns in the non-coding genome without requiring labels of ...
genomebiology.biomedcentral.com
Peter Koo
@pkoo562.bsky.social
· Jul 16
Peter Koo
@pkoo562.bsky.social
· Jul 16
Peter Koo
@pkoo562.bsky.social
· Jul 16
Peter Koo
@pkoo562.bsky.social
· Jul 16
Peter Koo
@pkoo562.bsky.social
· Jul 16
Peter Koo
@pkoo562.bsky.social
· Jul 16
Peter Koo
@pkoo562.bsky.social
· Jul 16
Peter Koo
@pkoo562.bsky.social
· Jul 16
Peter Koo
@pkoo562.bsky.social
· Jul 16
Peter Koo
@pkoo562.bsky.social
· Jul 16

Evaluating the representational power of pre-trained DNA language models for regulatory genomics - Genome Biology
Background The emergence of genomic language models (gLMs) offers an unsupervised approach to learning a wide diversity of cis-regulatory patterns in the non-coding genome without requiring labels of ...
genomebiology.biomedcentral.com