Samantha Graham
@samanthagraham.bsky.social
66 followers
58 following
13 posts
PhD student at UMN studying the microbiome | she/her | spgraham1.github.io
Posts
Media
Videos
Starter Packs
Reposted by Samantha Graham
Ran Blekhman
@blekhman.bsky.social
· Jun 18
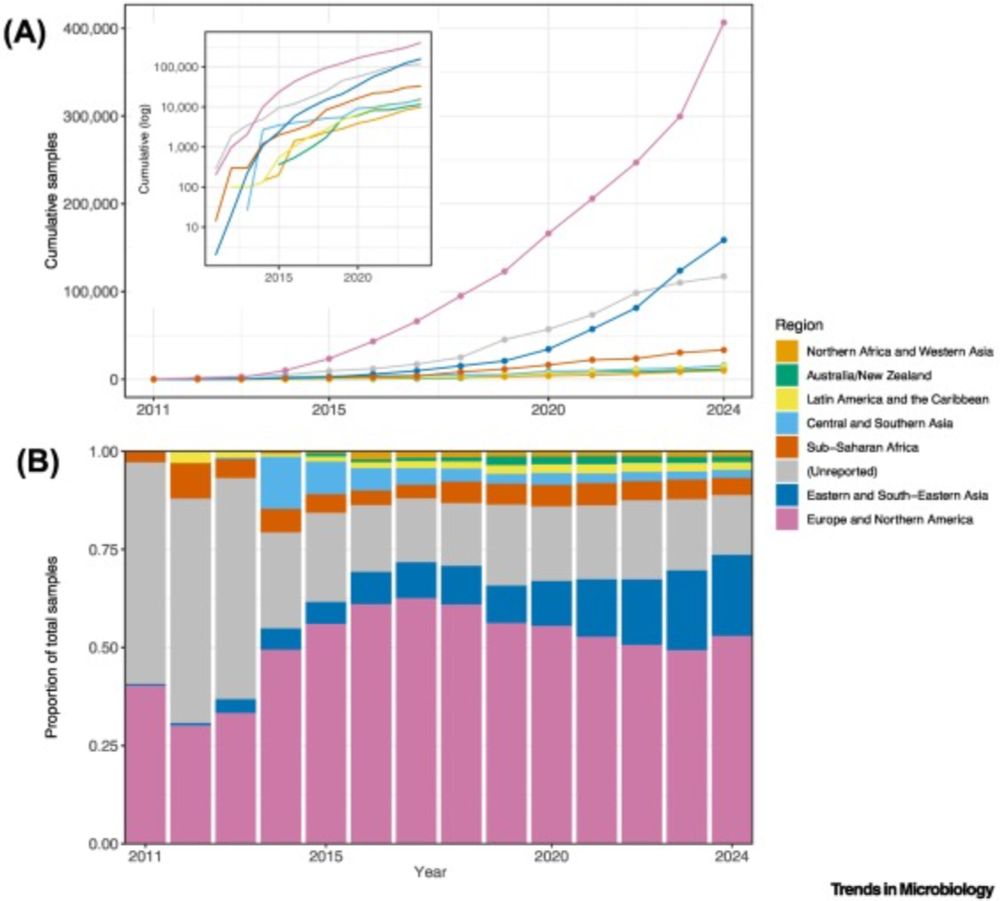
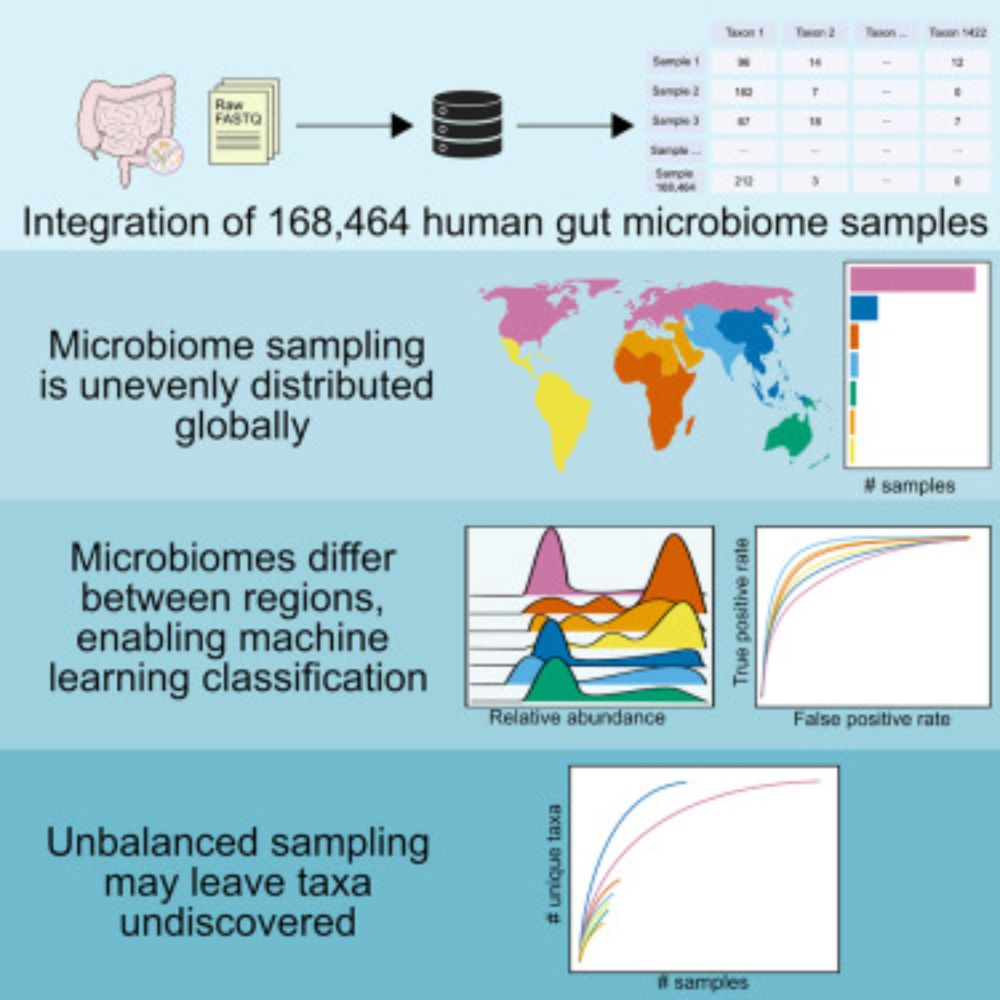
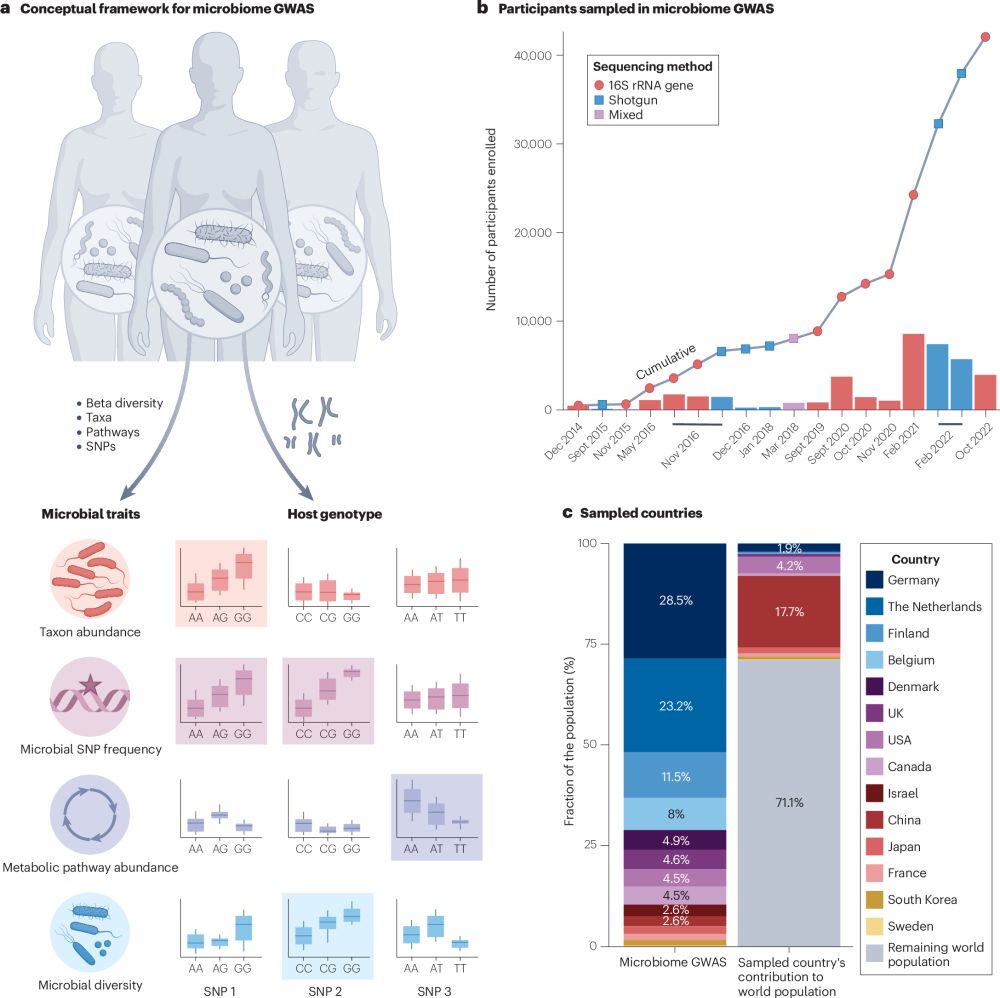
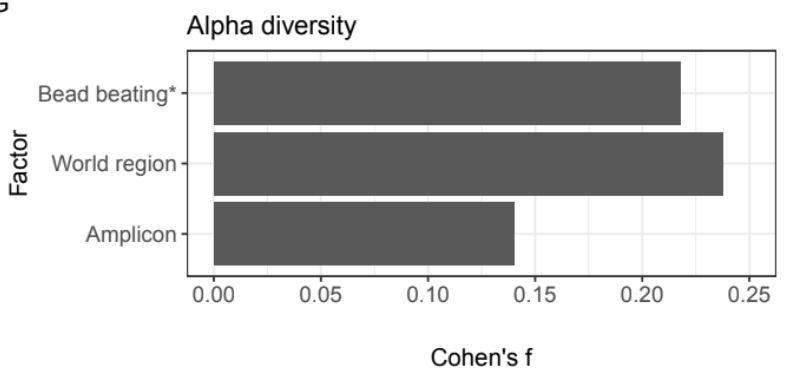
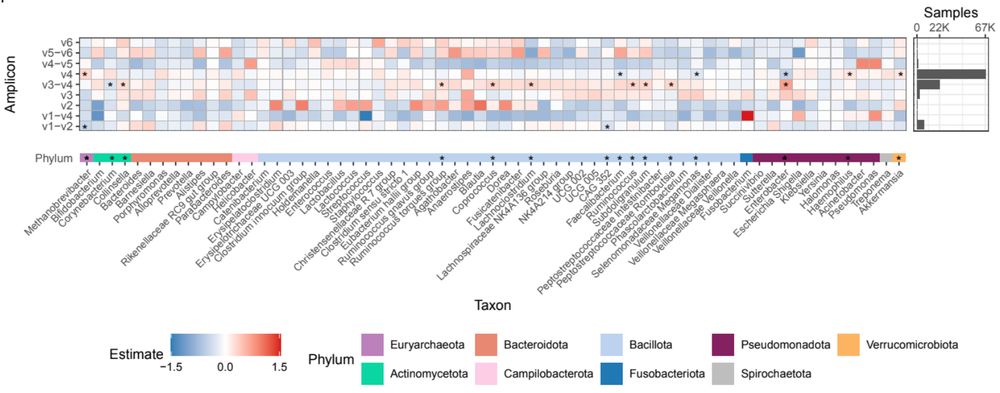
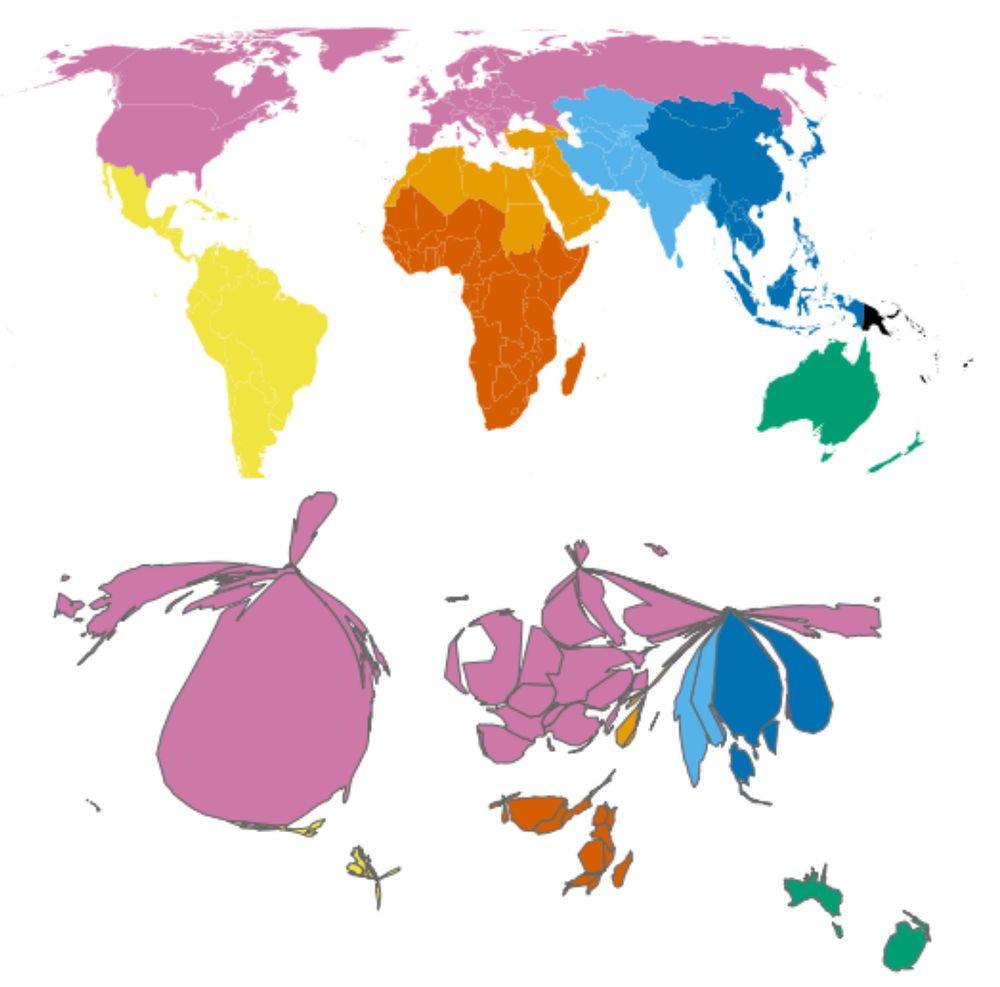
Analyzing human gut microbiome data from global populations: challenges and resources
Research on the human gut microbiome is expanding rapidly; yet, most published studies
focus on populations from high-income regions such as North America and Europe. Underrepresentation
of population...
www.cell.com
Reposted by Samantha Graham
Rich Abdill
@richabdill.com
· May 19

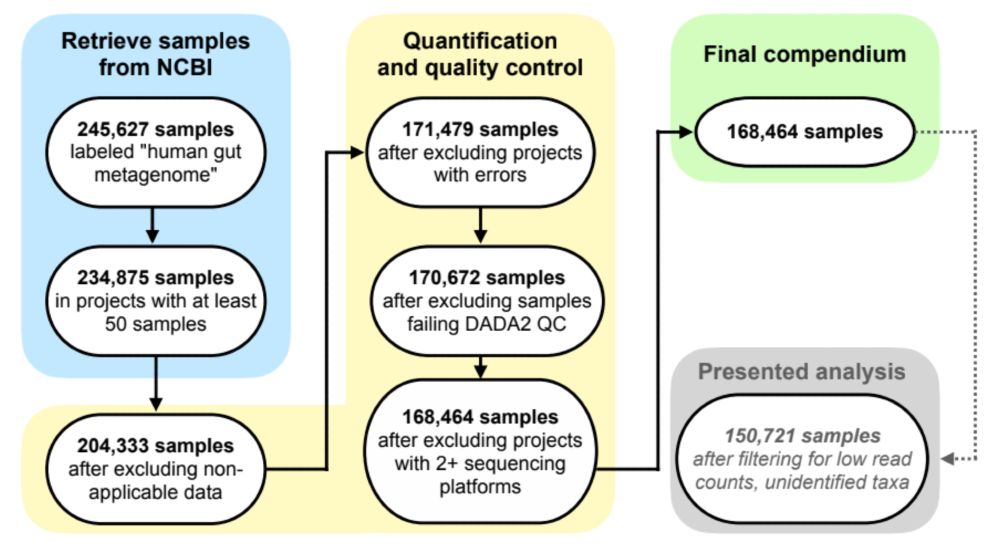
Compendium Manager: a tool for coordination of workflow management instances for bulk data processing in Python
Compendium Manager is a command-line tool written in Python to automate the provisioning, launch, and evaluation of bioinformatics pipelines. Although workflow management tools such as Snakemake and N...
arxiv.org
Reposted by Samantha Graham
Reposted by Samantha Graham
Reposted by Samantha Graham
Ran Blekhman
@blekhman.bsky.social
· Jan 24