Sergio Andreu-Sanchez
@seandreu.bsky.social
160 followers
610 following
16 posts
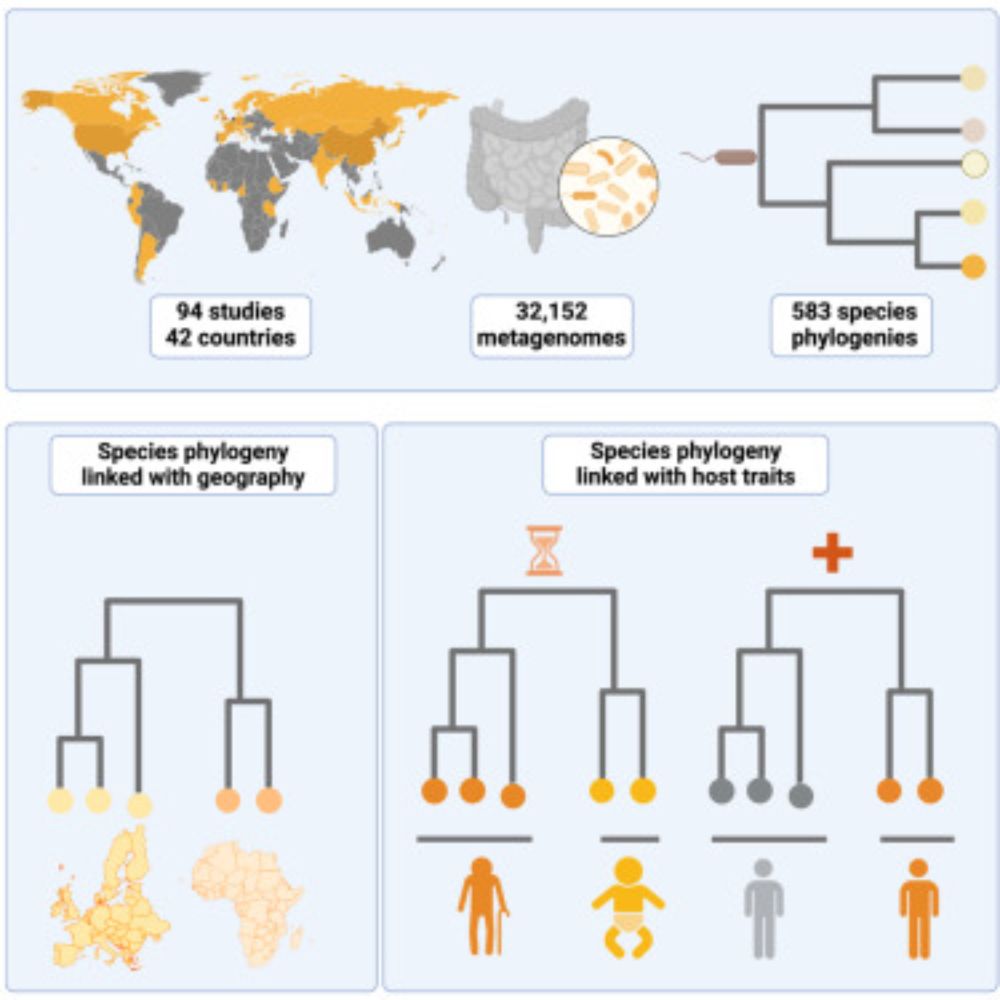
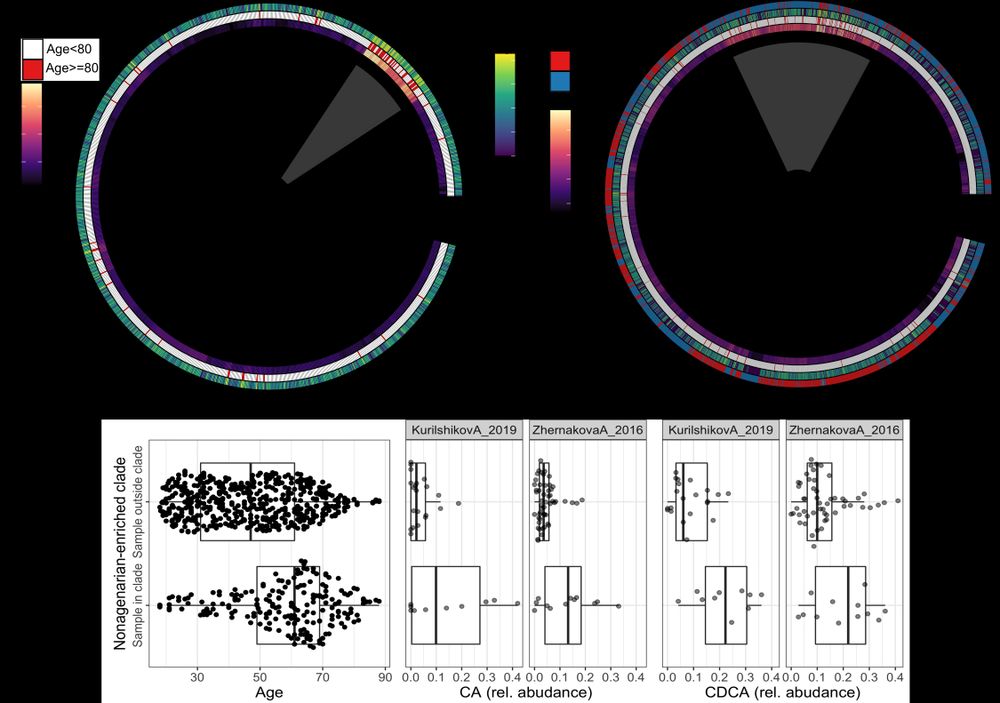
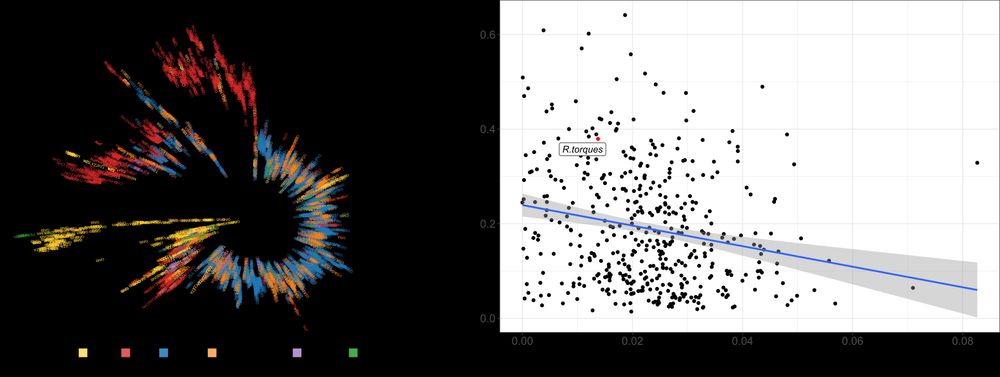
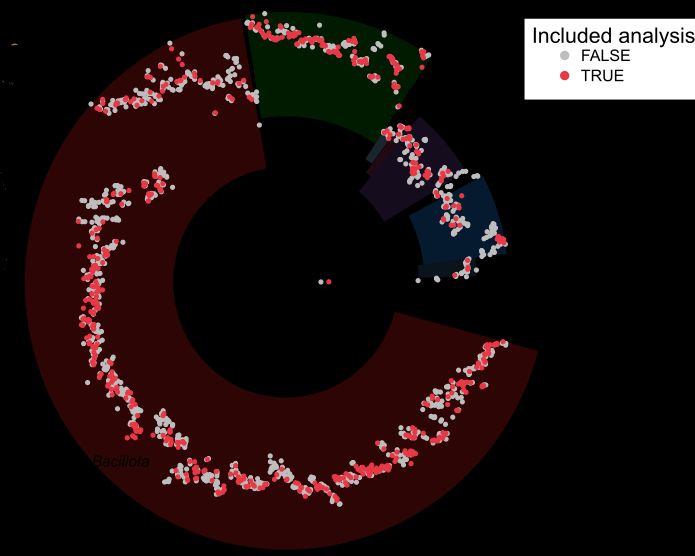
Postdoctoral researcher at @raeslab.org, former Groningen Microbiome Hub.
(meta)Genomics, bioinformatics
Posts
Media
Videos
Starter Packs
Reposted by Sergio Andreu-Sanchez
Reposted by Sergio Andreu-Sanchez
Reposted by Sergio Andreu-Sanchez
Reposted by Sergio Andreu-Sanchez
Reposted by Sergio Andreu-Sanchez
Sebastian Schmidt
@tsbschm.bsky.social
· Jun 27
Reposted by Sergio Andreu-Sanchez
Reposted by Sergio Andreu-Sanchez
Segata Lab
@cibiocm.bsky.social
· Apr 30
Reposted by Sergio Andreu-Sanchez
Rob Lanfear
@roblanfear.bsky.social
· May 27
Reposted by Sergio Andreu-Sanchez
Reposted by Sergio Andreu-Sanchez
Reposted by Sergio Andreu-Sanchez