
And we mean scratch: a blinded study.
TLDR: 26 days. And the binders work…and led to new cancer biology.
We’re coming for you AI….
chemrxiv.org/engage/chemr...

And we mean scratch: a blinded study.
TLDR: 26 days. And the binders work…and led to new cancer biology.
We’re coming for you AI….
chemrxiv.org/engage/chemr...

#ChemBio #ChemSky #DrugDiscovery

#ChemBio #ChemSky #DrugDiscovery
pubs.acs.org/doi/10.1021/...

pubs.acs.org/doi/10.1021/...
1/n

1/n
www.nature.com/articles/s41...
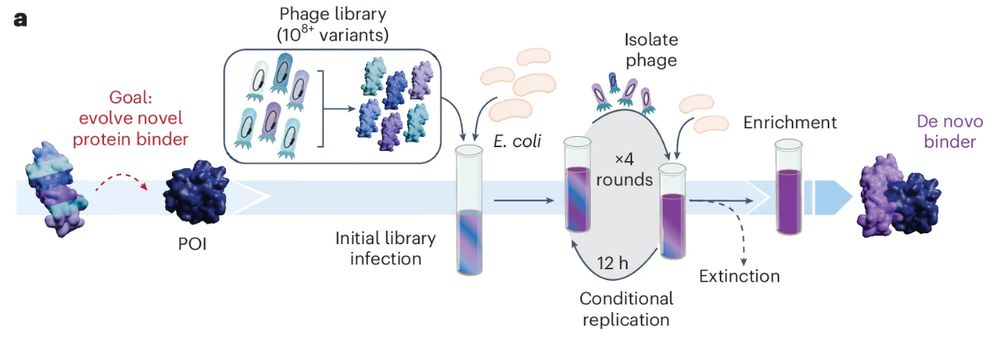
www.nature.com/articles/s41...
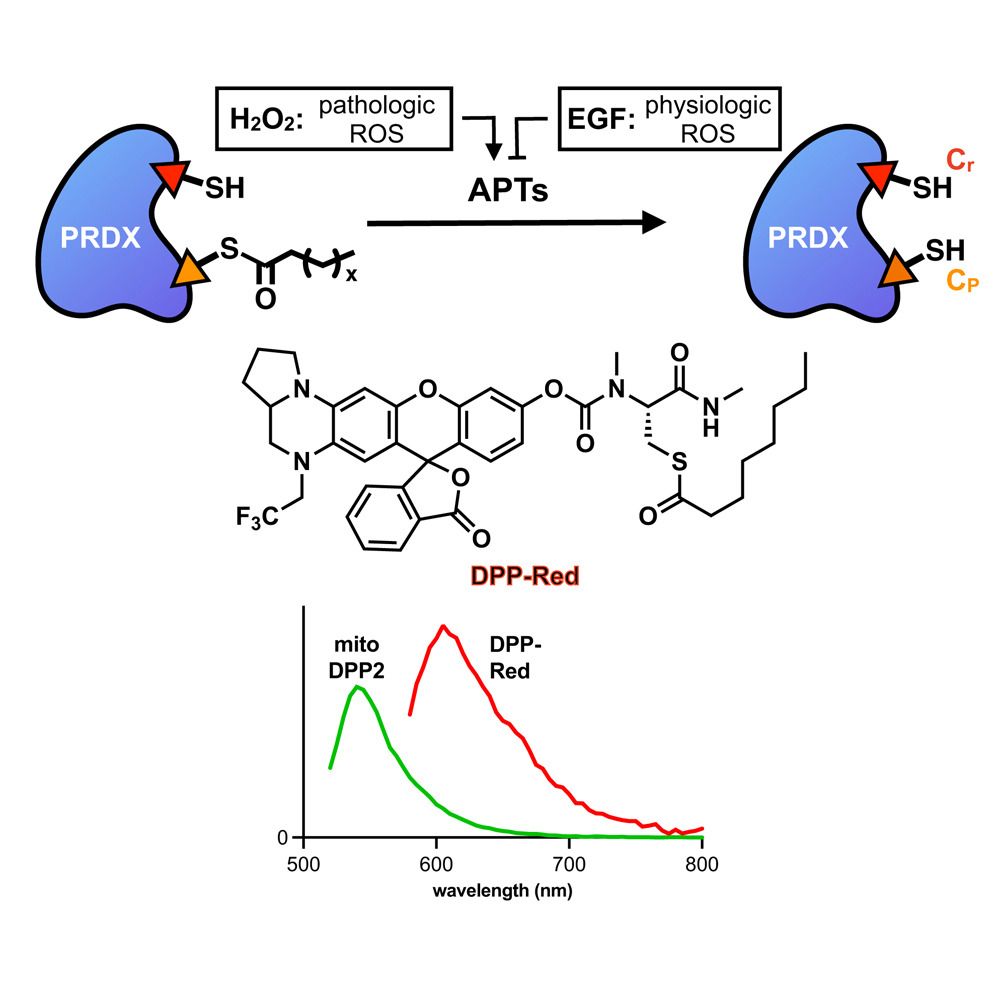
Congrats Tian Saara-Anne & @shubha-pani.bsky.social
authors.elsevier.com/a/1kg6b8jWWJ...
Congrats Tian Saara-Anne & @shubha-pani.bsky.social
authors.elsevier.com/a/1kg6b8jWWJ...
TLDR: PANCS-binders is fast (2 days), cheap (pennies), has extremely high fidelity (low false positive and negatives), and high-throughput.
1/n
#biosky

TLDR: PANCS-binders is fast (2 days), cheap (pennies), has extremely high fidelity (low false positive and negatives), and high-throughput.
1/n

