Sudarshan Pinglay
@sudpinglay.bsky.social
270 followers
200 following
45 posts
Scientist at UW Genome Sciences and the Seattle Hub for Synthetic Biology.
http://pinglay-lab.com/
synBio/genomics/soccer/heavy metal/food
Posts
Media
Videos
Starter Packs
Pinned
Sudarshan Pinglay
@sudpinglay.bsky.social
· Jan 31
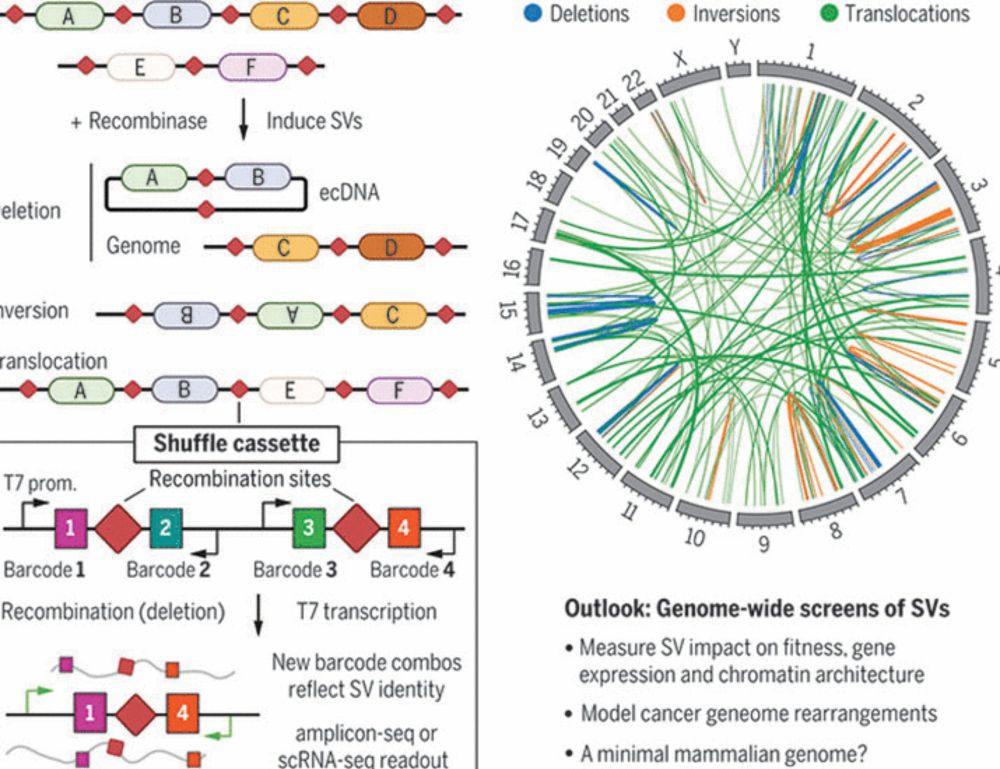
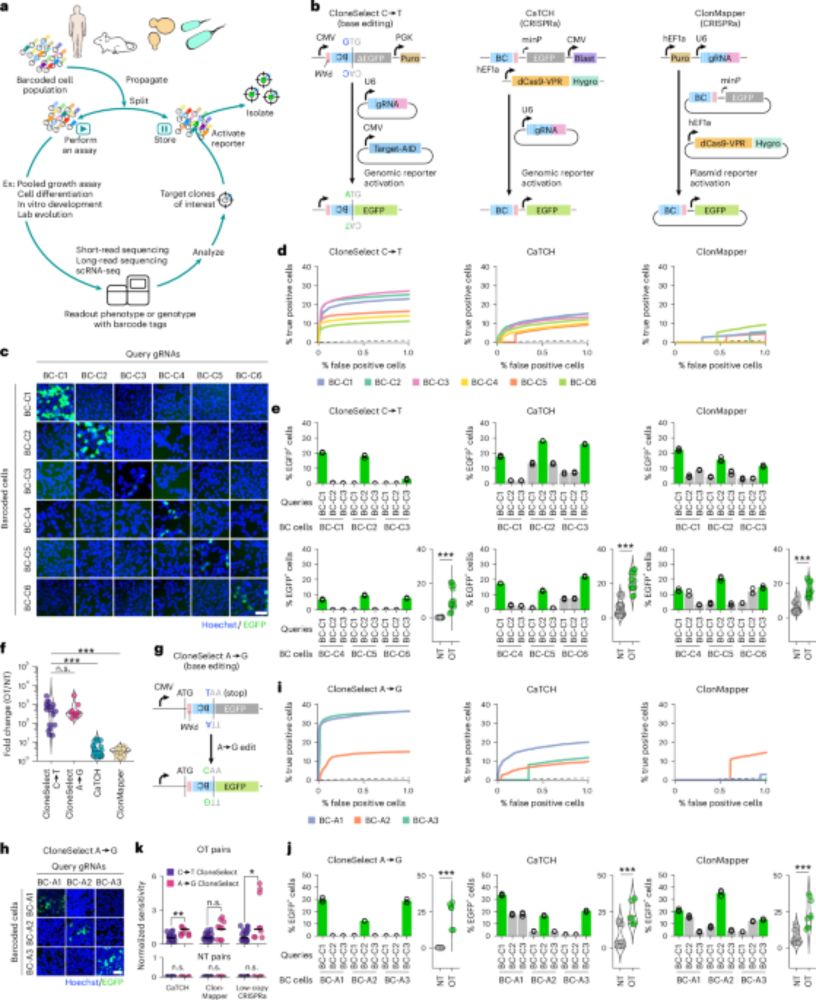
Multiplex generation and single-cell analysis of structural variants in mammalian genomes
Studying the functional consequences of structural variants (SVs) in mammalian genomes is challenging because (i) SVs arise much less commonly than single-nucleotide variants or small indels and (ii) ...
www.science.org
Reposted by Sudarshan Pinglay
Reposted by Sudarshan Pinglay
Abby Buchwalter
@abbybuch.bsky.social
· Jul 21

Lamin B1 and LAP2β resist cytoskeletal force to maintain lamin A/C meshwork organization and preserve nuclear integrity | Molecular Biology of the Cell
The nuclear lamins are extremely long-lived proteins in most cell types. As a consequence, lamin function cannot be effectively dissected with temporal precision using standard knock-down approaches. ...
www.molbiolcell.org
Sudarshan Pinglay
@sudpinglay.bsky.social
· Jul 14

A shotgun approach for highly multiplexed mammalian metabolic engineering
Mammalian metabolic engineering is critical to advancing basic biology, bioproduction, and cell therapy. However, as pathway complexity increases, so does the size of both the combinatorial design spa...
www.biorxiv.org
Sudarshan Pinglay
@sudpinglay.bsky.social
· Jul 14
Sudarshan Pinglay
@sudpinglay.bsky.social
· Jul 14

A shotgun approach for highly multiplexed mammalian metabolic engineering
Mammalian metabolic engineering is critical to advancing basic biology, bioproduction, and cell therapy. However, as pathway complexity increases, so does the size of both the combinatorial design spa...
doi.org
Sudarshan Pinglay
@sudpinglay.bsky.social
· Jul 14
Sudarshan Pinglay
@sudpinglay.bsky.social
· Jul 14
Sudarshan Pinglay
@sudpinglay.bsky.social
· Jul 14

A shotgun approach for highly multiplexed mammalian metabolic engineering
Mammalian metabolic engineering is critical to advancing basic biology, bioproduction, and cell therapy. However, as pathway complexity increases, so does the size of both the combinatorial design spa...
doi.org
Reposted by Sudarshan Pinglay
Reposted by Sudarshan Pinglay
Sudarshan Pinglay
@sudpinglay.bsky.social
· Apr 25
Sudarshan Pinglay
@sudpinglay.bsky.social
· Apr 11
Reposted by Sudarshan Pinglay
Dave Truong
@davidmtruong.bsky.social
· Apr 11
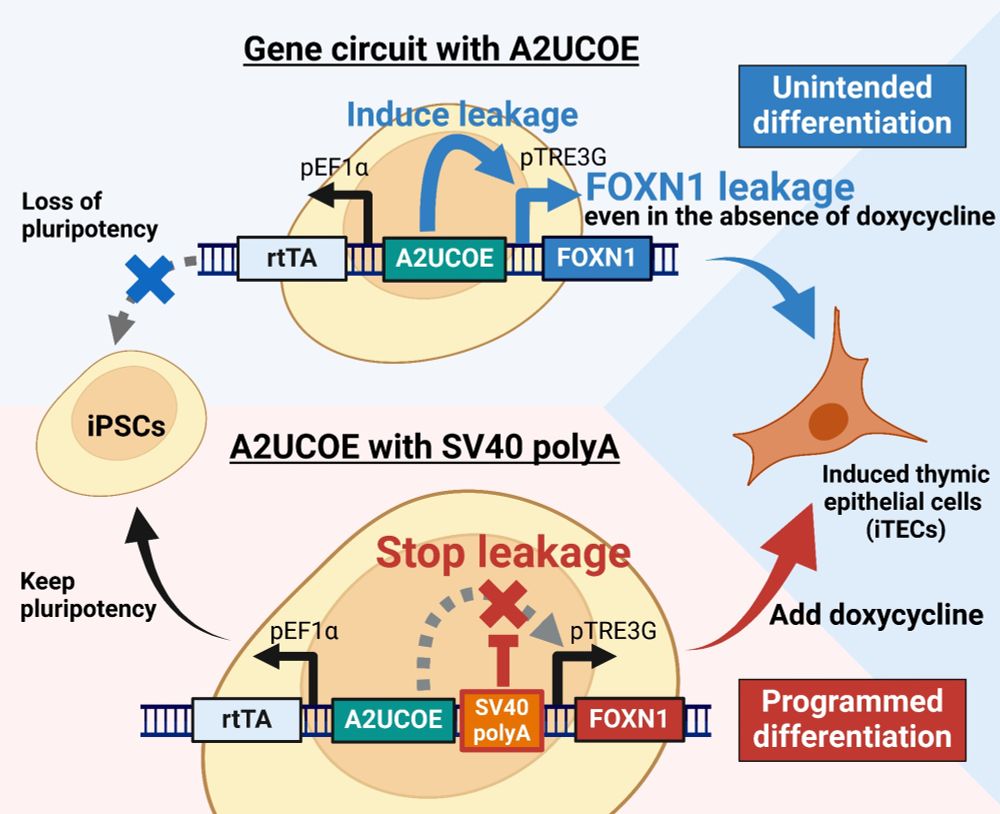
Termination sequence between an inducible promoter and ubiquitous chromatin opening element (UCOE) reduces gene expression leakage and silencing - Journal of Biological Engineering
Background Inducible gene expression circuits enable precise control over target gene activation and are widely used in direct reprogramming. However, their usability is often compromised by DNA methy...
tinyurl.com