Tami Gjorgjieva
@tamigj.bsky.social
140 followers
150 following
7 posts
Complex traits, gene regulation, ELSI (ethical, legal, and social implications of) genetics
PhD candidate with the Pritchard Lab @Stanford
Posts
Media
Videos
Starter Packs
Reposted by Tami Gjorgjieva
Tami Gjorgjieva
@tamigj.bsky.social
· Aug 9
Reposted by Tami Gjorgjieva
Reposted by Tami Gjorgjieva
Reposted by Tami Gjorgjieva
Katie Houlahan
@khoulahan.bsky.social
· Jan 8

Complex rearrangements fuel ER+ and HER2+ breast tumours - Nature
A study identifies three dominant genomic archetypes of breast cancer induced by discrete mutational processes, describing a continuum of genomic profiles and detailing the mechanisms underlying the p...
www.nature.com
Reposted by Tami Gjorgjieva
Jeff Spence
@jeffspence.github.io
· Dec 17

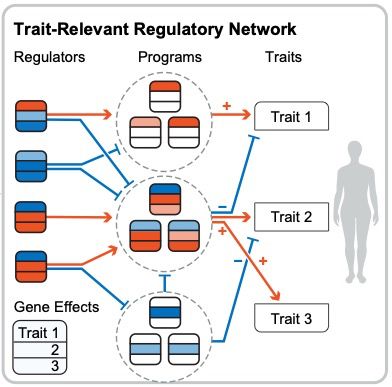
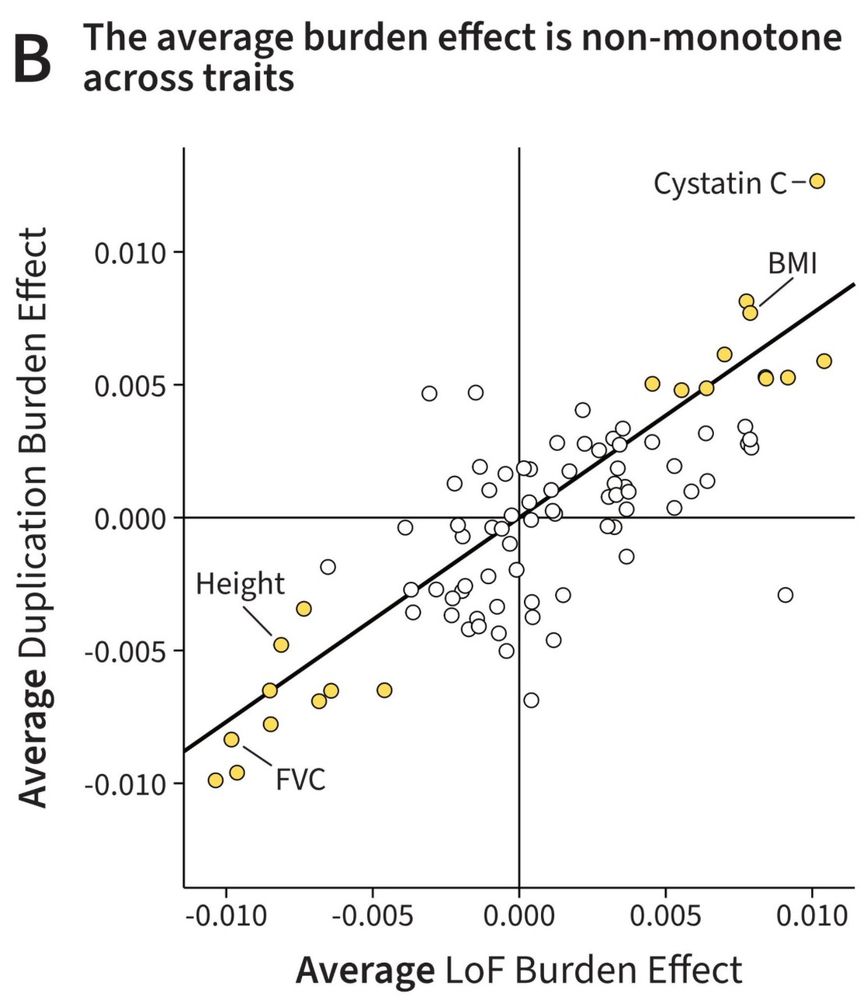
Specificity, length, and luck: How genes are prioritized by rare and common variant association studies
Standard genome-wide association studies (GWAS) and rare variant burden tests are essential tools for identifying trait-relevant genes. Although these methods are conceptually similar, we show by anal...
www.biorxiv.org
Reposted by Tami Gjorgjieva
Reposted by Tami Gjorgjieva
Reposted by Tami Gjorgjieva
Laura Helmuth
@laurahelmuth.bsky.social
· Nov 14