Brian Northan
@truenorth-ia.bsky.social
470 followers
390 following
47 posts
Contract Research and Development Engineer, Image Processing. XC skier. Stockade-athon 15 km Running Road Race Director.
Posts
Media
Videos
Starter Packs
Brian Northan
@truenorth-ia.bsky.social
· Aug 16
Reposted by Brian Northan
Reposted by Brian Northan
Reposted by Brian Northan
Reposted by Brian Northan
Reposted by Brian Northan
Brian Northan
@truenorth-ia.bsky.social
· Jul 15
Brian Northan
@truenorth-ia.bsky.social
· Jul 12
Brian Northan
@truenorth-ia.bsky.social
· Jul 12
Brian Northan
@truenorth-ia.bsky.social
· Jun 17
Reposted by Brian Northan
Reposted by Brian Northan
Reposted by Brian Northan
Reposted by Brian Northan
Dave Barry
@david-j-barry.bsky.social
· May 30
Reposted by Brian Northan
Brian Northan
@truenorth-ia.bsky.social
· May 27
Brian Northan
@truenorth-ia.bsky.social
· May 27

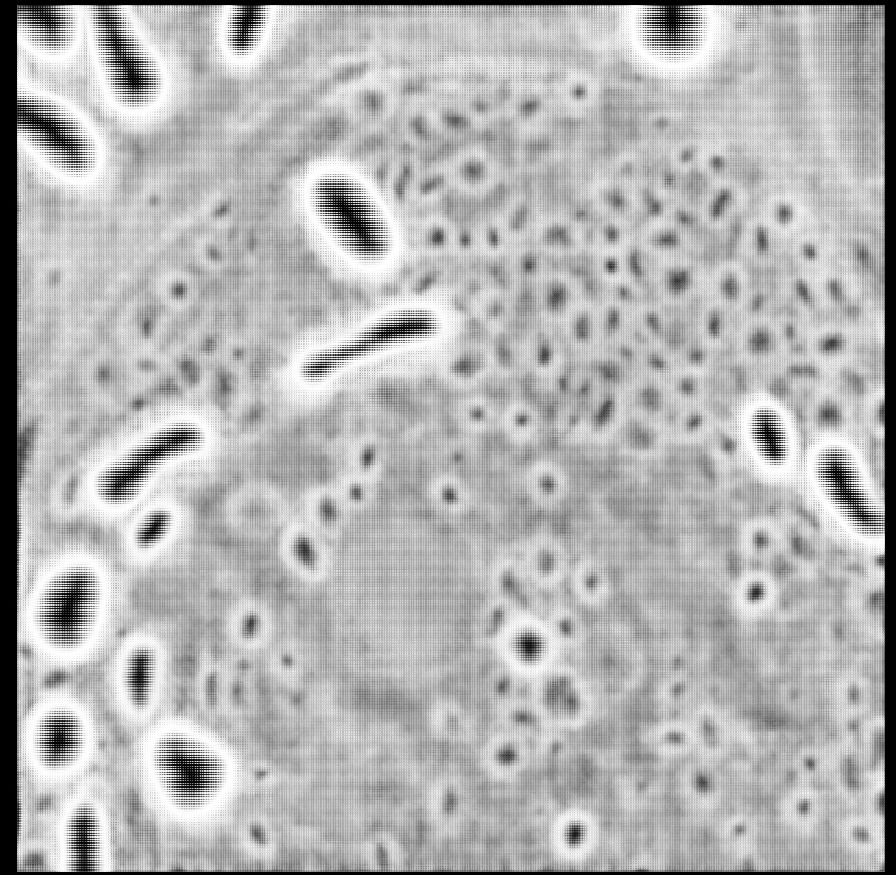
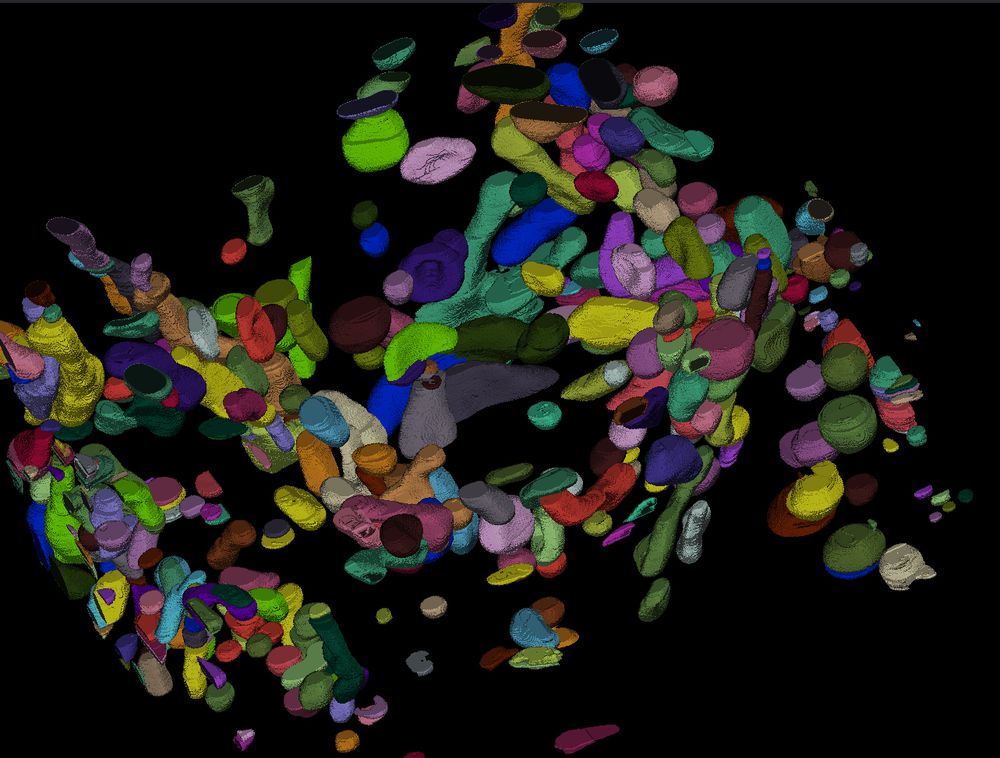
Are we seeing downstream effects of ambiguous labelling on new generalist models?
Below is a rather long post about some issues I’ve been seeing in generalist models ‘finding’ and ‘not finding’ objects, and my hypothesis as to why. Love to hear if others are seeing similar. The i...
forum.image.sc
Brian Northan
@truenorth-ia.bsky.social
· May 20
Brian Northan
@truenorth-ia.bsky.social
· May 20
Reposted by Brian Northan
Reposted by Brian Northan
Reposted by Brian Northan
Beth Cimini 🔬💻📊
@bethcimini.bsky.social
· May 15