Alumnus @borklab.bsky.social
Microbiome, microbial ecology & metagenomics.

I call this "The World according to Metagenomic Sampling Bias"
4/

I call this "The World according to Metagenomic Sampling Bias"
4/
We semi-manually annotated 305k publicly available metagenomes; n of samples per microntology term are shown in the plot.
3/

We semi-manually annotated 305k publicly available metagenomes; n of samples per microntology term are shown in the plot.
3/
And in @nature.com no less...
www.nature.com/articles/s41...

And in @nature.com no less...
www.nature.com/articles/s41...
We have received thoughtful (and critical) feedback on the initial version and look forward to receiving more!

We have received thoughtful (and critical) feedback on the initial version and look forward to receiving more!

While deeper sampled habitats had lower ρ (as expected), there were interesting deviations from the trend: hot springs harbour more small clades than expected, the oral cavity is disproportionately dominated by large clades.

While deeper sampled habitats had lower ρ (as expected), there were interesting deviations from the trend: hot springs harbour more small clades than expected, the oral cavity is disproportionately dominated by large clades.
x axis in plot is 'clade size' (i.e., n of subclades)


x axis in plot is 'clade size' (i.e., n of subclades)


Give it a try, and feedback is always welcome!

Give it a try, and feedback is always welcome!



Guess the researcher based on G Scholar citation stats:
Guess the researcher based on G Scholar citation stats:
But also quite literally. I once put this as a biosketch into a talk:

But also quite literally. I once put this as a biosketch into a talk:
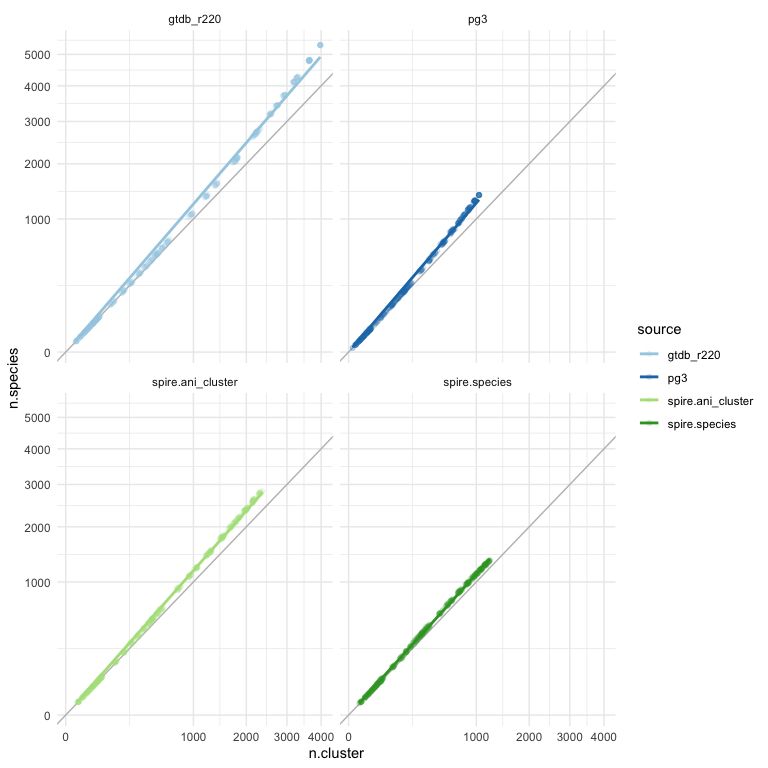
NB: `spire.species` are GTDB classifications (only) for SPIRE MAGs, `spire.ani` is just 95% clusters of SPIRE MAGs as a "species" equiv.
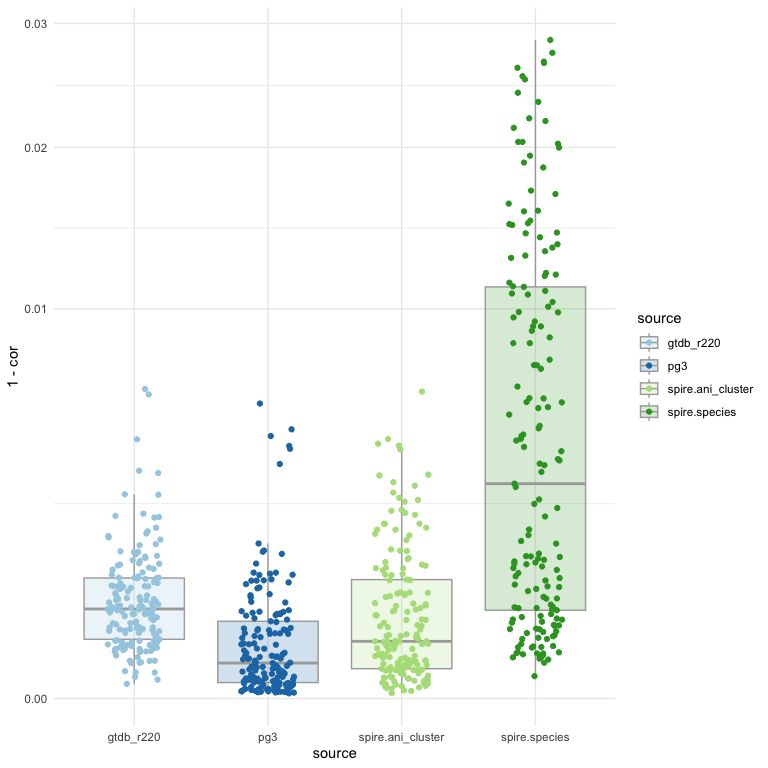
NB: `spire.species` are GTDB classifications (only) for SPIRE MAGs, `spire.ani` is just 95% clusters of SPIRE MAGs as a "species" equiv.
For genera, we find that >3 novel genera are discoverable per each recognized genus in the reference.
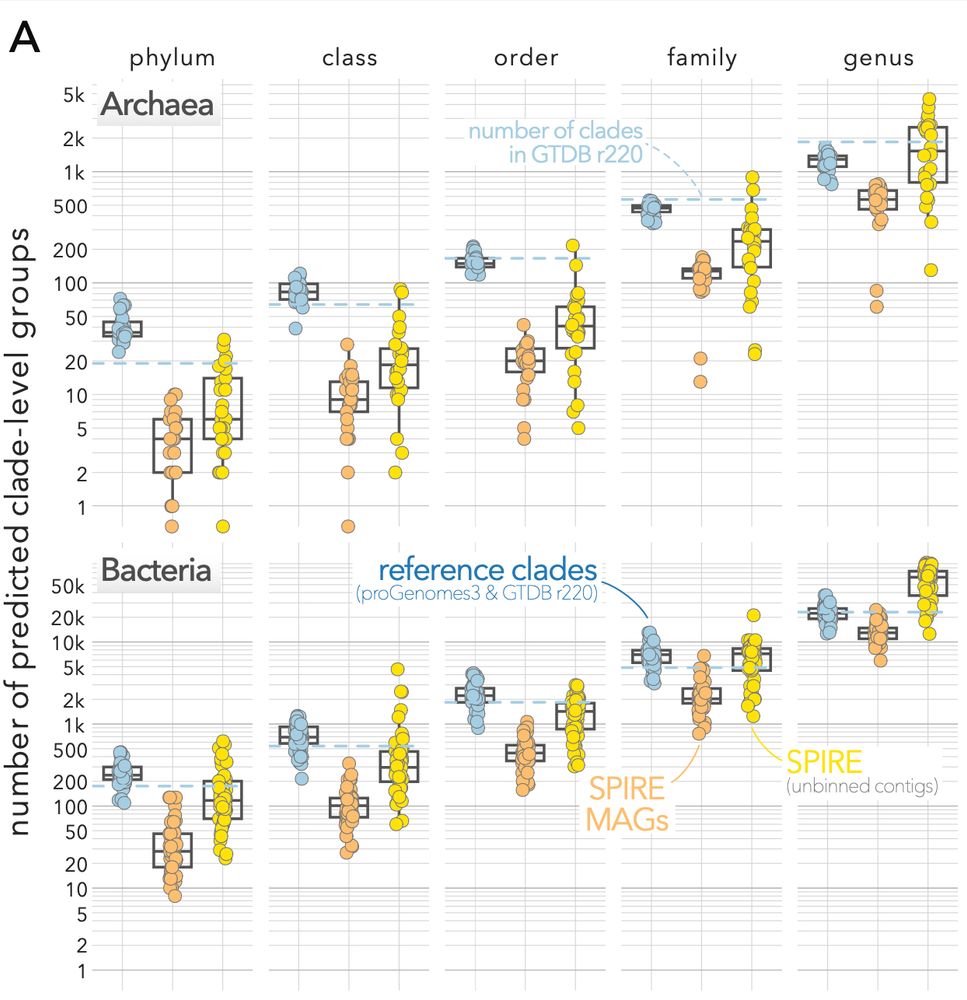
For genera, we find that >3 novel genera are discoverable per each recognized genus in the reference.
(dots in the plot below indicate phyla and families)
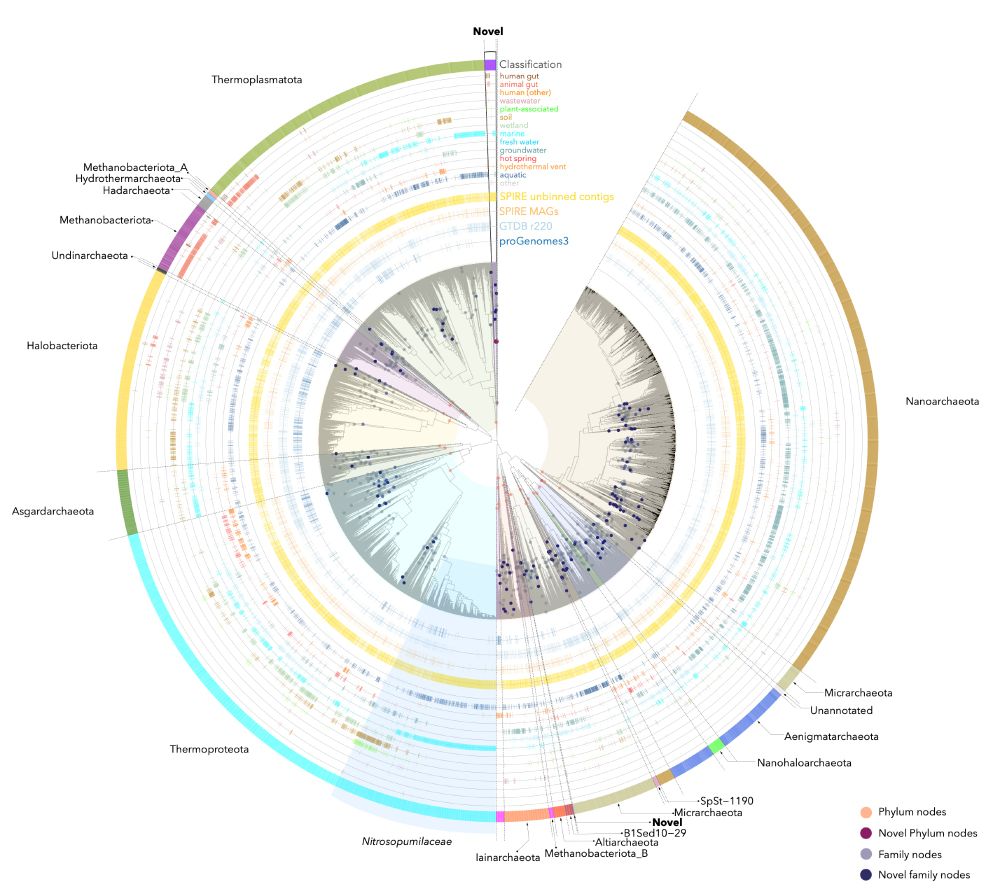
(dots in the plot below indicate phyla and families)
Most unbinned species species lurk among soil, aquatic & wetland samples.
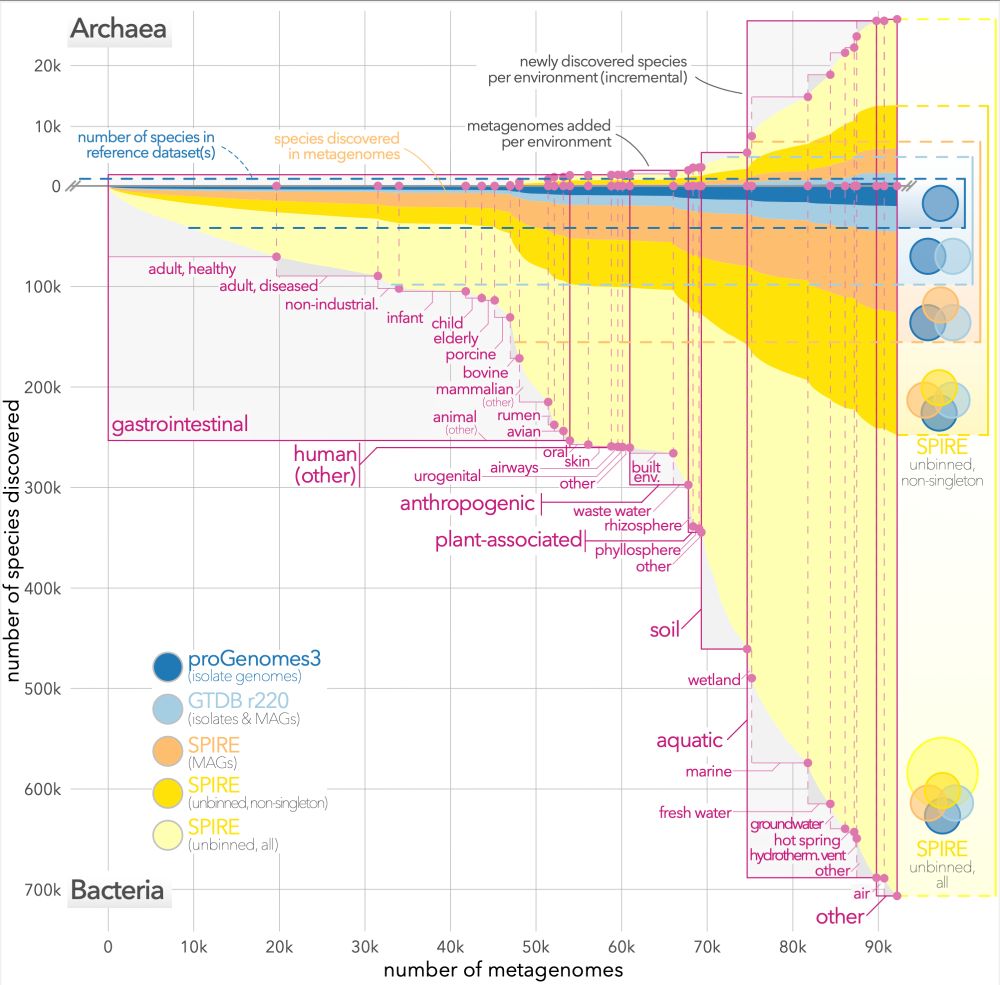
Most unbinned species species lurk among soil, aquatic & wetland samples.
Few habitats show signes of saturation (no new sp. added as more samples come in, α≤0). Most, in particular soils & aquatic envs, remain in full discovery swing.
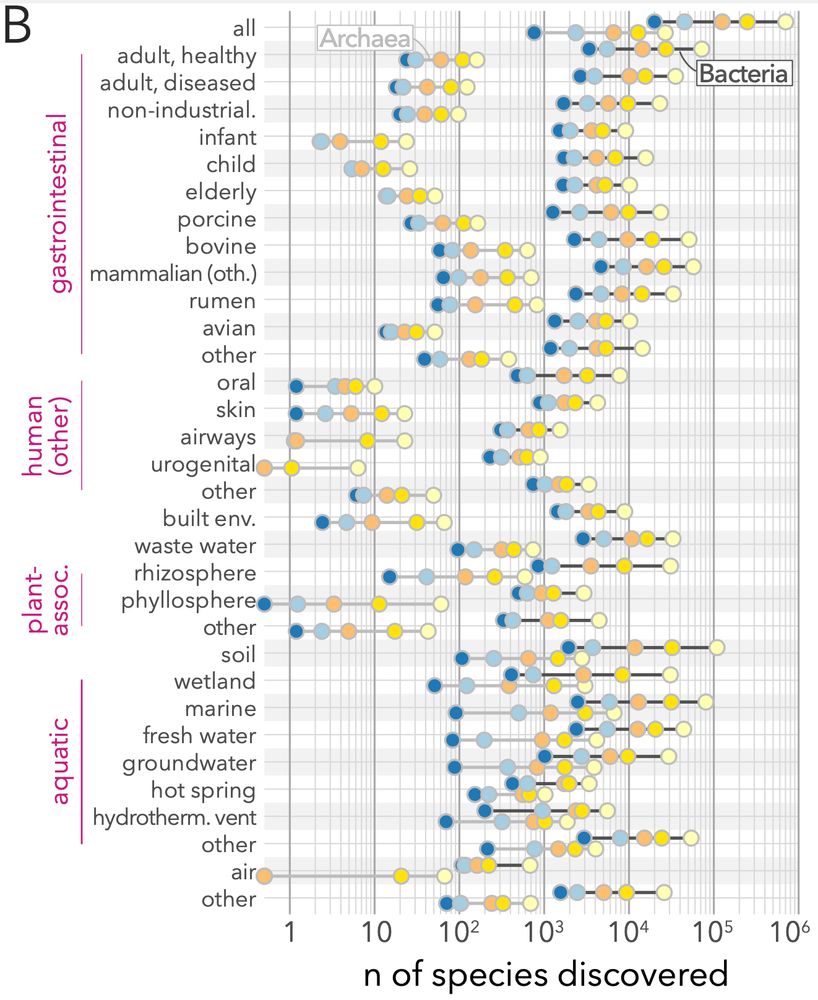
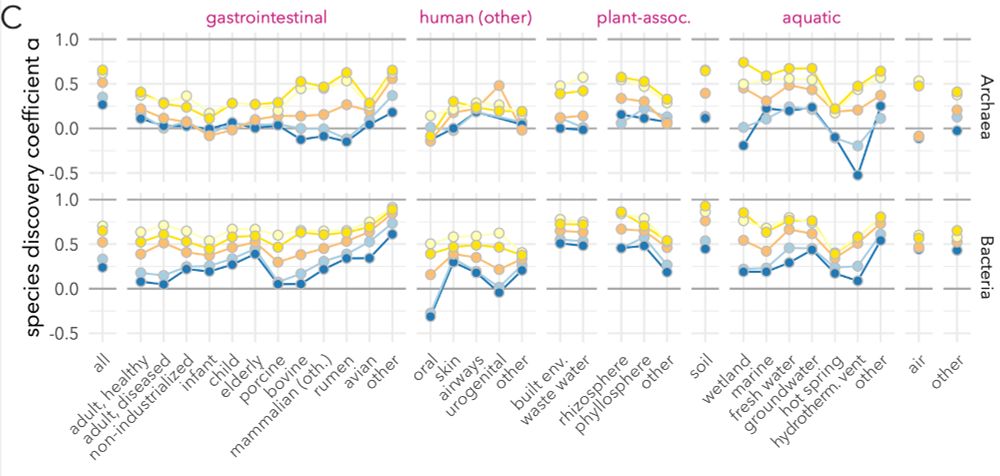
Few habitats show signes of saturation (no new sp. added as more samples come in, α≤0). Most, in particular soils & aquatic envs, remain in full discovery swing.
We estimate that ~702k bacterial and ~27k archaeal species are "discoverable" in total in these contigs.

We estimate that ~702k bacterial and ~27k archaeal species are "discoverable" in total in these contigs.




