Mark Williams single molecule biophysics lab
@williamslabneu.bsky.social
170 followers
230 following
3 posts
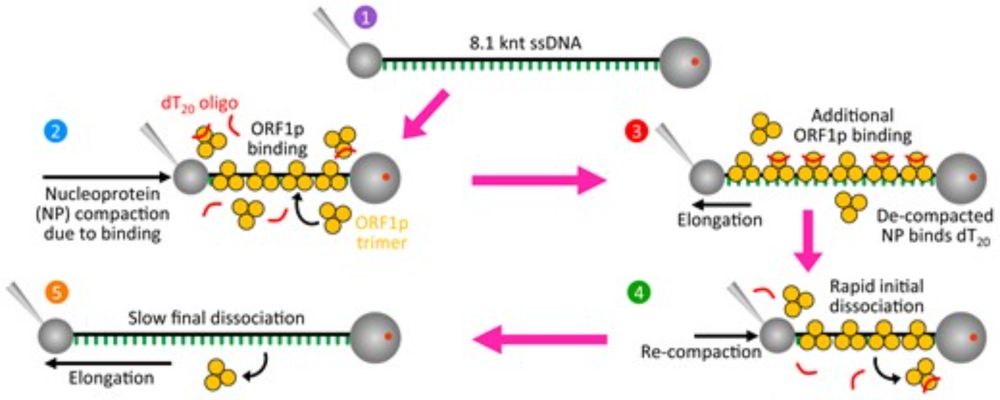
Optical tweezers | AFM | correlated fluorescence and force | nucleic acid-protein interactions | SSBs | chromatin stability | nucleosome chaperones | retrovirus, coronavirus, retrotransposon replication
https://williamslab.sites.northeastern.edu
Posts
Media
Videos
Starter Packs