Yan Hu
@yanhu97.bsky.social
78 followers
92 following
18 posts
Postdoctoral Researcher in the Srivastava Lab at the Gladstone Institutes. Buenrostro Lab Alumni. Interested in gene regulation, computational biology, aging, and human diseases.
Posts
Media
Videos
Starter Packs
Yan Hu
@yanhu97.bsky.social
· Jan 27
Reposted by Yan Hu
Vijay Ramani
@vram142.bsky.social
· Jan 24

Pervasive and programmed nucleosome distortion patterns on single mammalian chromatin fibers
We present a genome-scale method to map the single-molecule co-occupancy of structurally distinct nucleosomes, subnucleosomes, and other protein-DNA interactions via long-read high-resolution adenine ...
www.biorxiv.org
Yan Hu
@yanhu97.bsky.social
· Jan 27
Yan Hu
@yanhu97.bsky.social
· Jan 23
Yan Hu
@yanhu97.bsky.social
· Jan 23
Yan Hu
@yanhu97.bsky.social
· Jan 23
Yan Hu
@yanhu97.bsky.social
· Jan 23
Yan Hu
@yanhu97.bsky.social
· Jan 23
Yan Hu
@yanhu97.bsky.social
· Jan 23
Yan Hu
@yanhu97.bsky.social
· Jan 23
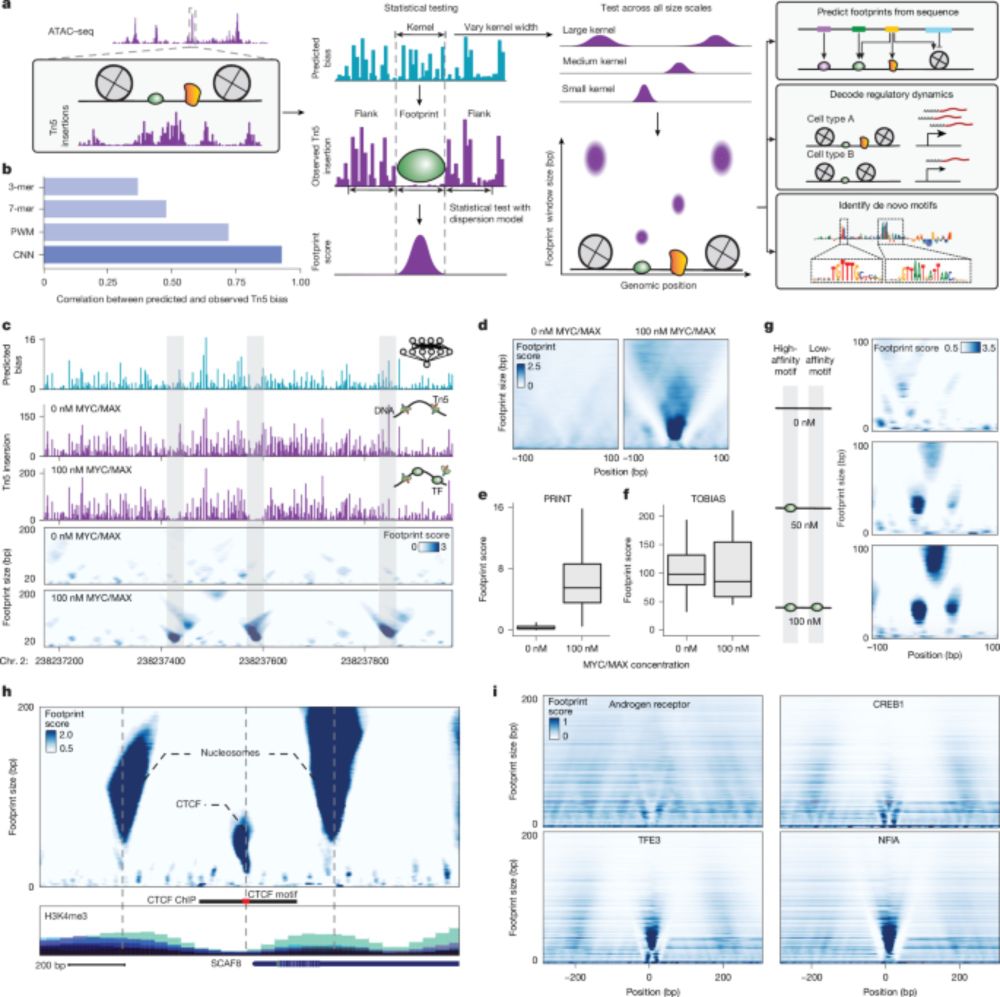
Multiscale footprints reveal the organization of cis-regulatory elements - Nature
We developed PRINT, a computational method that identifies footprints of DNA–protein interactions from bulk and single-cell chromatin accessibility data across multiple scales of protein size.
www.nature.com
Reposted by Yan Hu
Anshul Kundaje
@anshulkundaje.bsky.social
· Dec 25









