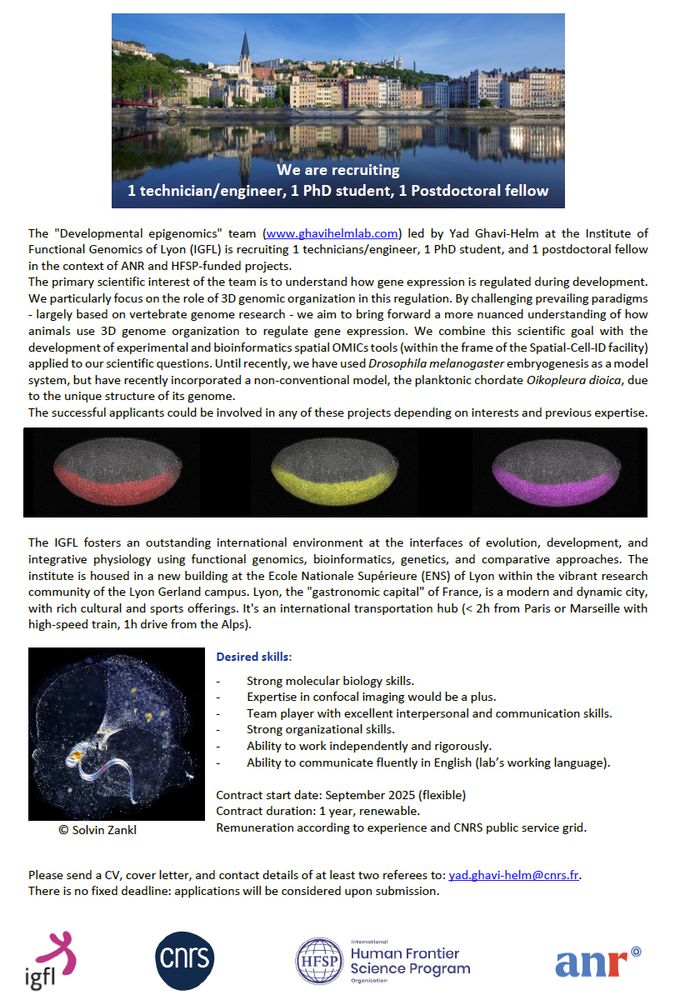
Yad Ghavi-Helm
@yghavi.bsky.social
1.5K followers
670 following
33 posts
Researcher @CNRS, PI @igflyon.bsky.social, Scientific coordinator #spatialOMICs facility @Spatial_Cell_ID facility @ENSdeLyon #enhancer #transcription #development #3Dgenome #Drosophila
Posts
Media
Videos
Starter Packs
Yad Ghavi-Helm
@yghavi.bsky.social
· Jul 28
Yad Ghavi-Helm
@yghavi.bsky.social
· Jul 24
Yad Ghavi-Helm
@yghavi.bsky.social
· Jul 24
Reposted by Yad Ghavi-Helm
Yad Ghavi-Helm
@yghavi.bsky.social
· Jul 23
Yad Ghavi-Helm
@yghavi.bsky.social
· Jul 23
Yad Ghavi-Helm
@yghavi.bsky.social
· Jul 23
Yad Ghavi-Helm
@yghavi.bsky.social
· Jul 23
Reposted by Yad Ghavi-Helm
Yad Ghavi-Helm
@yghavi.bsky.social
· Apr 8
Yad Ghavi-Helm
@yghavi.bsky.social
· Apr 2
Yad Ghavi-Helm
@yghavi.bsky.social
· Nov 20
Yad Ghavi-Helm
@yghavi.bsky.social
· Nov 19
Yad Ghavi-Helm
@yghavi.bsky.social
· Nov 18
Yad Ghavi-Helm
@yghavi.bsky.social
· Nov 18
Yad Ghavi-Helm
@yghavi.bsky.social
· Nov 18

Spatial reconstruction of single-cell enhancer activity in a multicellular organism
Enhancers play an essential role in developmental processes by orchestrating the spatial and temporal regulation of gene expression. However, mapping the location of these regulatory elements in the g...
tinyurl.com
Yad Ghavi-Helm
@yghavi.bsky.social
· Nov 18
Yad Ghavi-Helm
@yghavi.bsky.social
· Nov 18
Yad Ghavi-Helm
@yghavi.bsky.social
· Nov 18