@zhoujt.bsky.social
14 followers
16 following
11 posts
Science Fellow @ Arc Institute; Computation Biology, Genomics, Single Cell
Posts
Media
Videos
Starter Packs
Pinned
Reposted
zhoujt.bsky.social
@zhoujt.bsky.social
· Mar 25
zhoujt.bsky.social
@zhoujt.bsky.social
· Mar 25
zhoujt.bsky.social
@zhoujt.bsky.social
· Mar 25

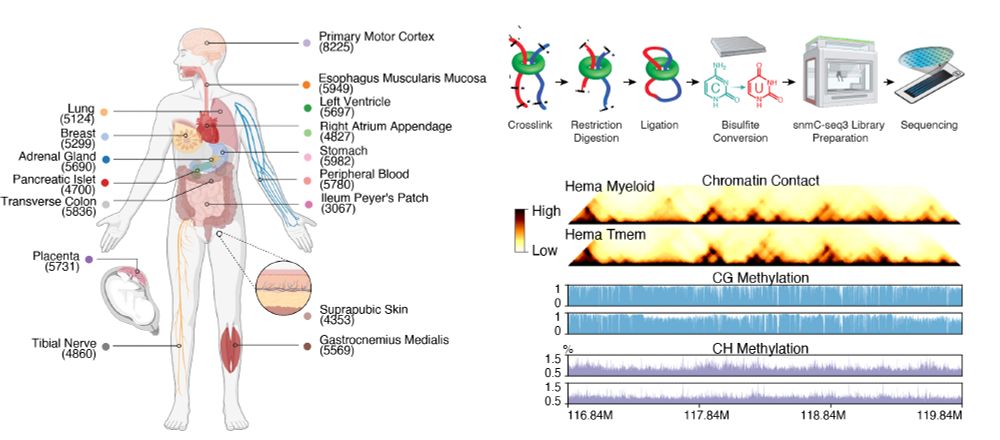
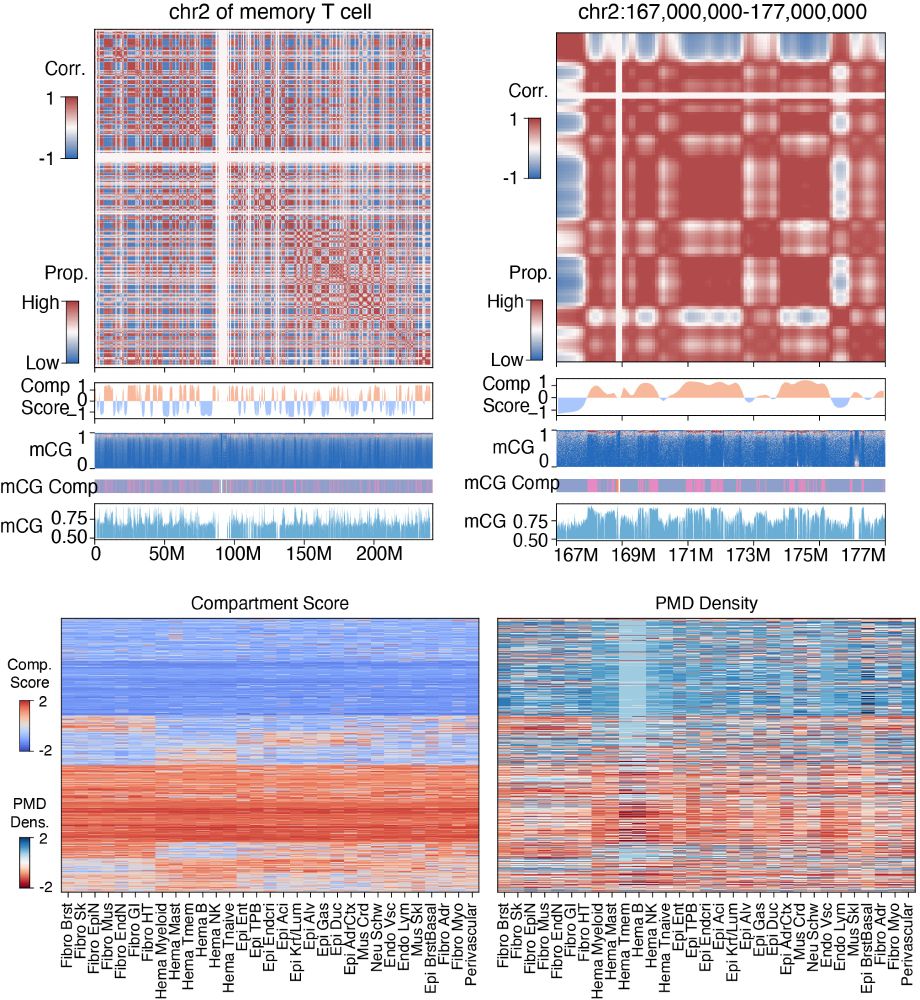
Human Body Single-Cell Atlas of 3D Genome Organization and DNA Methylation
Higher-order chromatin structure and DNA methylation are critical for gene regulation, but how these vary across the human body remains unclear. We performed multi-omic profiling of 3D genome structur...
www.biorxiv.org