[6/6]
[6/6]
I’m not quite sure I understand the question - maybe you could elaborate? The major benefit, however, of DySTrack is that you can correct/follow on the fly during acquisition rather than doing anything post!
I’m not quite sure I understand the question - maybe you could elaborate? The major benefit, however, of DySTrack is that you can correct/follow on the fly during acquisition rather than doing anything post!
Open-source, documented, and ready for feedback & contributions.
Check out the GitHub repo here:
github.com/WhoIsJack/Dy...
[5/6]

Open-source, documented, and ready for feedback & contributions.
Check out the GitHub repo here:
github.com/WhoIsJack/Dy...
[5/6]
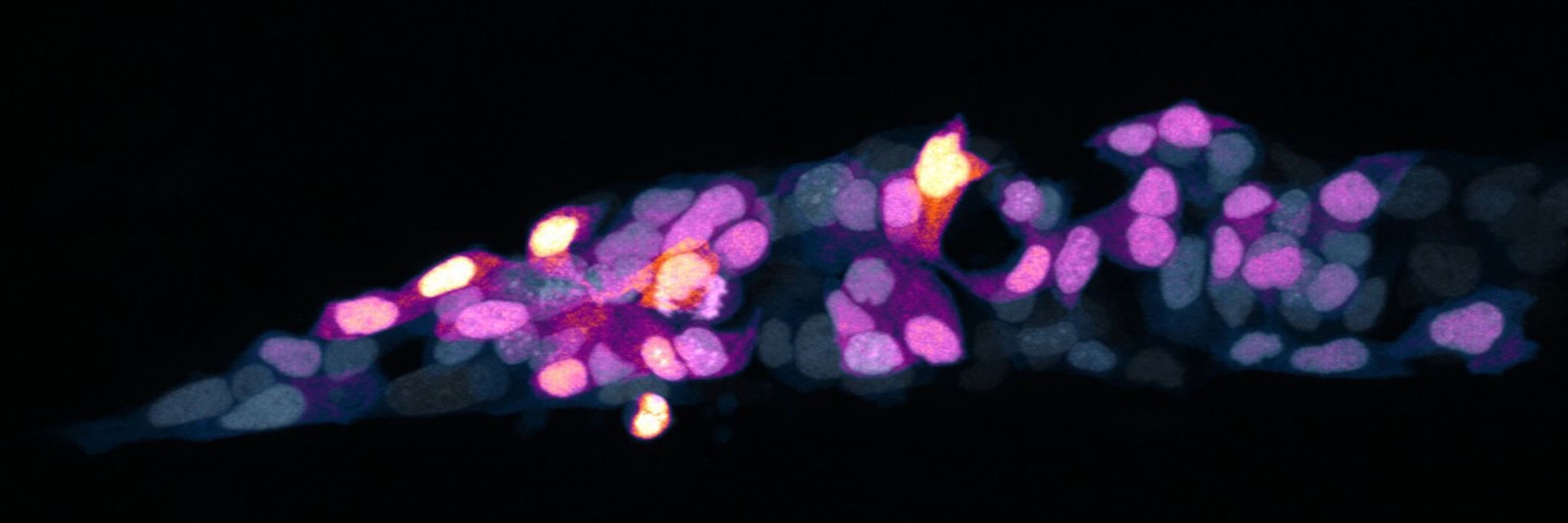
🐟 migrating zebrafish pLLP
🐟 zebrafish neuromast drift correction
🐣 chick Hensen’s node (pictured)
All easy to adapt to your own system.
[4/6]
🐟 migrating zebrafish pLLP
🐟 zebrafish neuromast drift correction
🐣 chick Hensen’s node (pictured)
All easy to adapt to your own system.
[4/6]
Your microscope can follow moving or drifting samples on its own with no vendor lock-in, no rewriting control software and no massive frameworks to learn. We provide integration for Zeiss LSM880, LSM980 and Nikon scopes that have JOBS.
[3/6]
Your microscope can follow moving or drifting samples on its own with no vendor lock-in, no rewriting control software and no massive frameworks to learn. We provide integration for Zeiss LSM880, LSM980 and Nikon scopes that have JOBS.
[3/6]
DySTrack lets you keep using Zeiss/Nikon software for acquisition while running any Python image analysis you want in the loop.
It watches prescans -> runs your analysis -> updates stage coords automatically.
[2/6]

DySTrack lets you keep using Zeiss/Nikon software for acquisition while running any Python image analysis you want in the loop.
It watches prescans -> runs your analysis -> updates stage coords automatically.
[2/6]

