
Prof. and Chair of Human Genetics at U. of Utah.
https://www.genetics.utah.edu/
http://quinlanlab.org

meetings.cshl.edu/meetings.asp...

meetings.cshl.edu/meetings.asp...
First authors: @saracarioscia.bsky.social & @aabiddanda.github.io

First authors: @saracarioscia.bsky.social & @aabiddanda.github.io
www.biorxiv.org/content/10.6...
www.biorxiv.org/content/10.6...

1/n
Move & resize the ranges to see how that affects bedtools operations like merge and intersect in real time!
Move & resize the ranges to see how that affects bedtools operations like merge and intersect in real time!

'In addition to Bhattacharya, the letter was sent to U.S. Health and Human Services Secretary Robert F. Kennedy Jr. and members of Congress who oversee NIH'
www.forbes.com/sites/michae...

'In addition to Bhattacharya, the letter was sent to U.S. Health and Human Services Secretary Robert F. Kennedy Jr. and members of Congress who oversee NIH'
www.forbes.com/sites/michae...

- We are seeking nominations bu June 1.
- September 18-19, 2025
- Please share with the star postdocs that you know.
docs.google.com/forms/d/e/1F...

- We are seeking nominations bu June 1.
- September 18-19, 2025
- Please share with the star postdocs that you know.
docs.google.com/forms/d/e/1F...
meetings.cshl.edu/meetings.asp...

meetings.cshl.edu/meetings.asp...
Check out the database resource, as STRchive "streamlines TR variant interpretation at disease-associated loci."
strchive.org
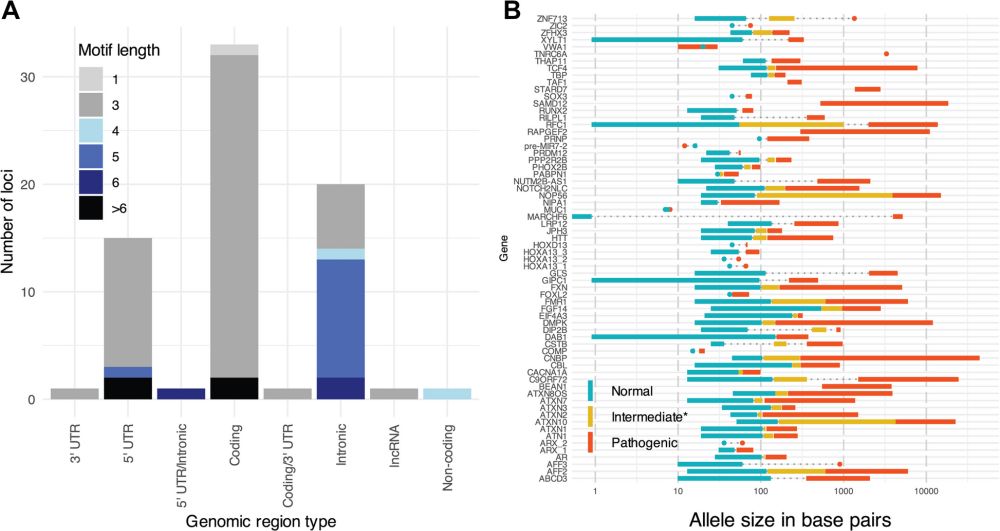
Check out the database resource, as STRchive "streamlines TR variant interpretation at disease-associated loci."
strchive.org
www.statnews.com/2025/03/17/t...

www.statnews.com/2025/03/17/t...
www.statnews.com/2025/03/14/n...

www.statnews.com/2025/03/14/n...
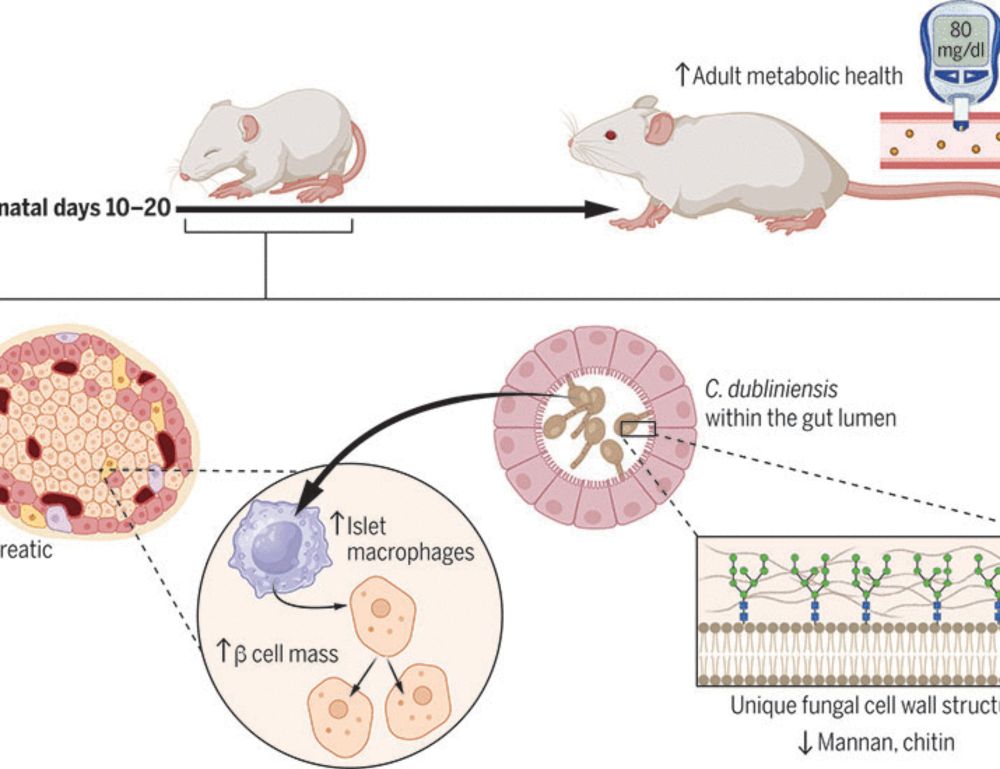
academic.oup.com/bioinformati...

academic.oup.com/bioinformati...
academic.oup.com/hropen/advan...

academic.oup.com/hropen/advan...




