https://www.genzentrum.uni-muenchen.de/research-groups/beckmann/index.html
rdcu.be/eRmJl
#ribosome #cryoEM #LMU #JHMI
rdcu.be/eRmJl
#ribosome #cryoEM #LMU #JHMI
#ERCSyG #GeneCenter #LMU #UniGraz #CNRS #JohnsHopkins

#ERCSyG #GeneCenter #LMU #UniGraz #CNRS #JohnsHopkins
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Cryo-EM & functional data by Roland Beckmann and coworkers show how Bcs1 alternates between different states during its translocation activity.
www.embopress.org/doi/full/10....
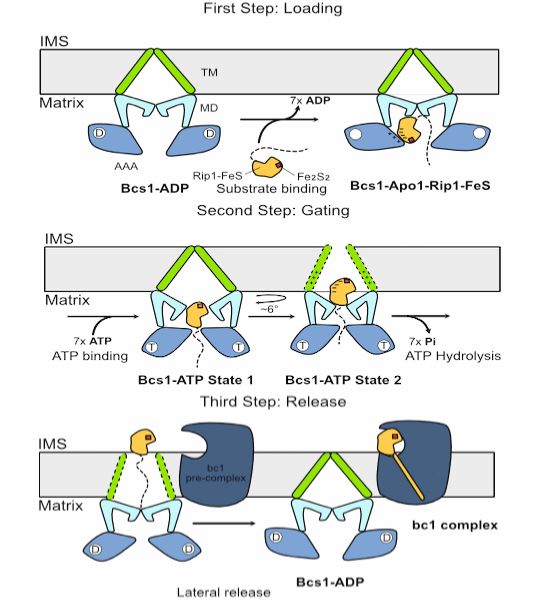
Cryo-EM & functional data by Roland Beckmann and coworkers show how Bcs1 alternates between different states during its translocation activity.
www.embopress.org/doi/full/10....

