
👨💻 Incoming assistant professor @University of Utah
🔗 Personal website: https://boyi-guo.com/
👤 Pronounce: he/him

The #GUOmics lab will focus on developing biologically informed machine learning methods to decode spatial and population-scale omics data. Look forward to exciting colabs!

Very excited how spatial transcriptomics contributes enlightening mediating effect of genetic risk variants.
Some cool work is brewing!
www.nature.com/articles/s41...

Very excited how spatial transcriptomics contributes enlightening mediating effect of genetic risk variants.
Some cool work is brewing!
www.nature.com/articles/s41...
Check our journal club session summary of the pre-print version by @lahuuki.bsky.social
The new features @boyiguo.bsky.social + @mictott.bsky.social added in this revised manuscript enhance the use cases for #SpotSweeper: 👀 #VisiumHD data!
youtu.be/f72XcXmtkfw?...
Check our journal club session summary of the pre-print version by @lahuuki.bsky.social
The new features @boyiguo.bsky.social + @mictott.bsky.social added in this revised manuscript enhance the use cases for #SpotSweeper: 👀 #VisiumHD data!
youtu.be/f72XcXmtkfw?...
Grateful for the opportunity to work together with @martinowk.bsky.social and @lieberinstitute.bsky.social, investigating the molecular pathology of neuropsychiatric disorder, coming soon! 😉
Applying this method to our QC workflow has substantially improved many new ST datasets generated across diverse regions/tissues in the human 🧠
We introduce SpotSweeper, the first spatially-aware QC methods for spatial transcriptomics.
📰 Paper : nature.com/articles/s41...
💻 Code: github.com/MicTott/Spot...
📈 Website: mictott.github.io/SpotSweeper/
🧵👇
Grateful for the opportunity to work together with @martinowk.bsky.social and @lieberinstitute.bsky.social, investigating the molecular pathology of neuropsychiatric disorder, coming soon! 😉
Applying this method to our QC workflow has substantially improved many new ST datasets generated across diverse regions/tissues in the human 🧠
We introduce SpotSweeper, the first spatially-aware QC methods for spatial transcriptomics.
📰 Paper : nature.com/articles/s41...
💻 Code: github.com/MicTott/Spot...
📈 Website: mictott.github.io/SpotSweeper/
🧵👇
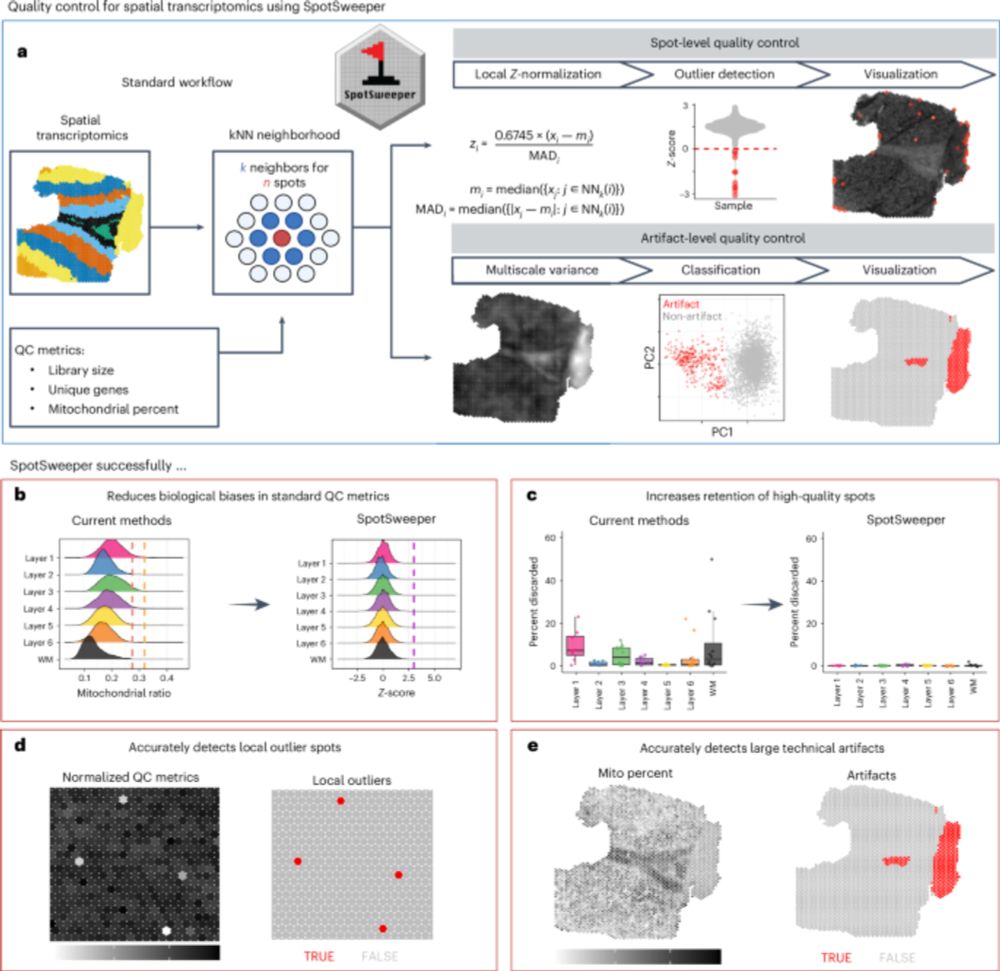
Applying this method to our QC workflow has substantially improved many new ST datasets generated across diverse regions/tissues in the human 🧠
The #GUOmics lab will focus on developing biologically informed machine learning methods to decode spatial and population-scale omics data. Look forward to exciting colabs!

The #GUOmics lab will focus on developing biologically informed machine learning methods to decode spatial and population-scale omics data. Look forward to exciting colabs!
SpotSweeper is developed to combat implicit selection bias.
Led by fellow rising star @mictott.bsky.social and collab with amazing mentor @stephaniehicks.bsky.social
👉: bsky.app/profile/mict...

SpotSweeper is developed to combat implicit selection bias.
Led by fellow rising star @mictott.bsky.social and collab with amazing mentor @stephaniehicks.bsky.social
👉: bsky.app/profile/mict...
www.nature.com/articles/s41...
Enjoyed reading the article, and reflect on my own writing workflow.
Plan to include in my lab handbook as resources for trainees.
#PDL#writing#comp-lab

www.nature.com/articles/s41...
Enjoyed reading the article, and reflect on my own writing workflow.
Plan to include in my lab handbook as resources for trainees.
#PDL#writing#comp-lab
💡 idea is to extend mutual nearest neighbors for
#spatial data. We call it spatial mutual nearest neighbors (spatialMNN) 😄
Thank you @haowen-zhou.bsky.social @pratibha-panwar.bsky.social who led this work! 👏 🧬🖥️🧪
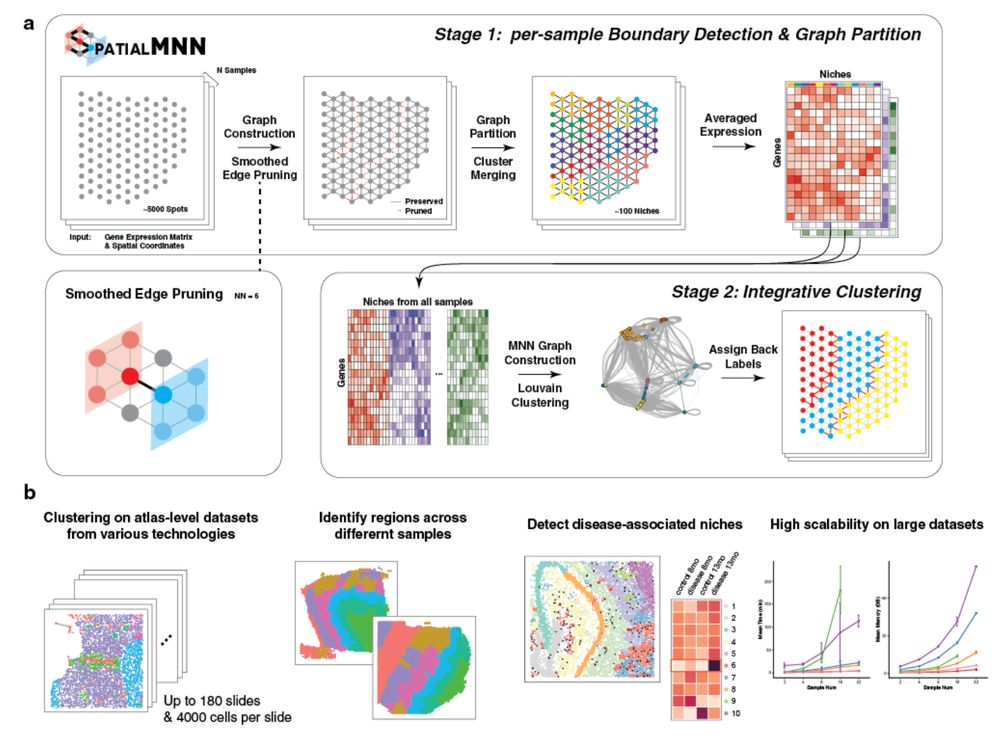
💡 idea is to extend mutual nearest neighbors for
#spatial data. We call it spatial mutual nearest neighbors (spatialMNN) 😄
Thank you @haowen-zhou.bsky.social @pratibha-panwar.bsky.social who led this work! 👏 🧬🖥️🧪
#ProudPI #stats 🖥️🧬🧪🧠📈
Preprint: www.biorxiv.org/content/10.1...
Software: www.bioconductor.org/packages/spoon


