umassmed.edu/kelchlab
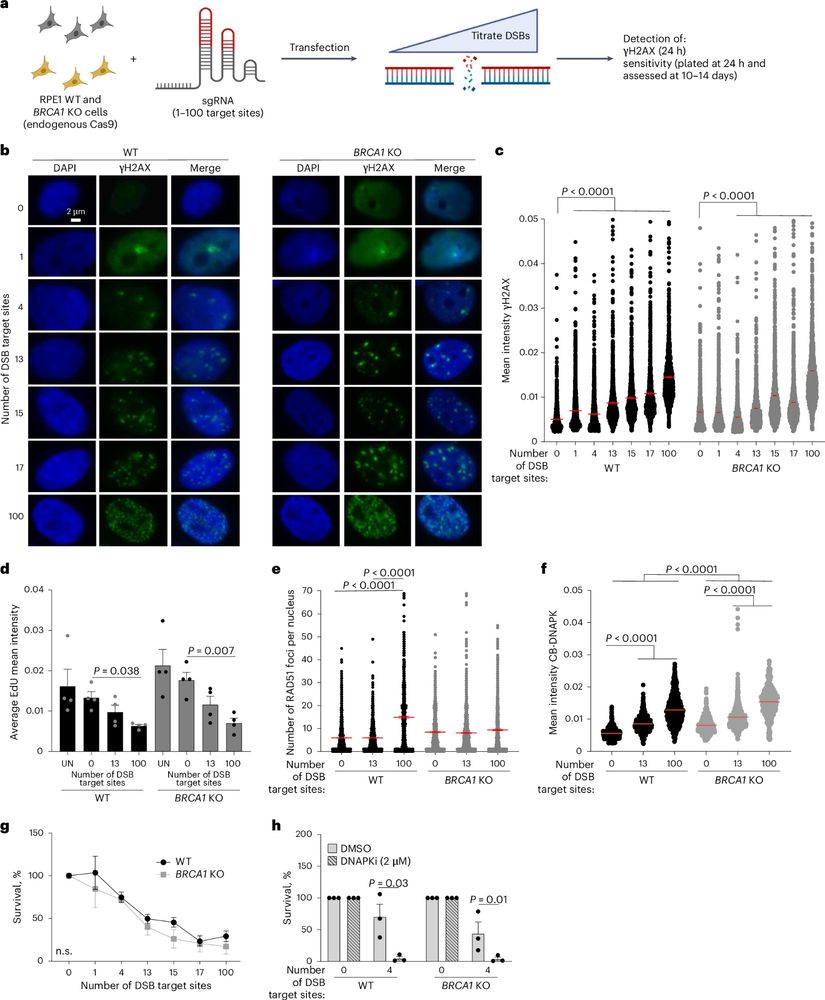
doi.org/10.1101/2025...

doi.org/10.1101/2025...
www.sciencedirect.com/science/arti...
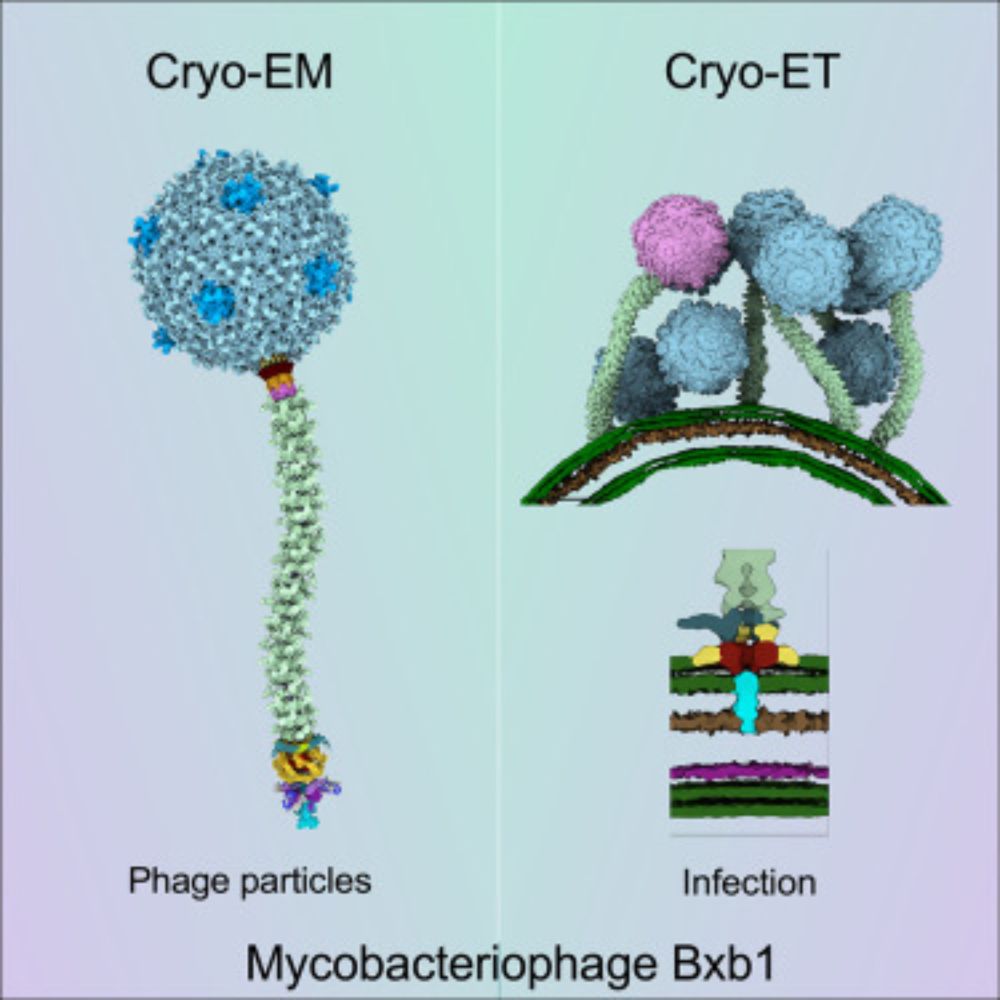
www.sciencedirect.com/science/arti...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
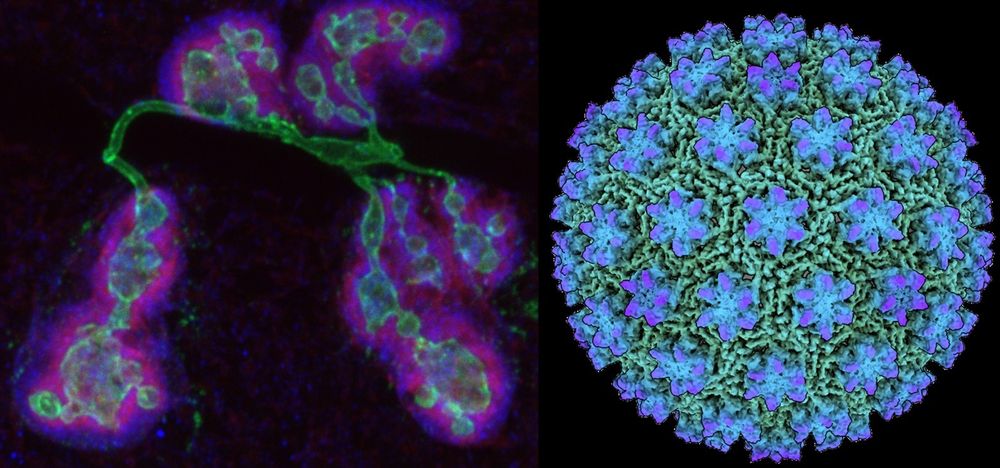
Did animals co-opt viral gag for intercellular (trans synaptic) communication? Or did viruses co-opt retrotransposon gags that animals had previously co-opted for intercellular use?

Did animals co-opt viral gag for intercellular (trans synaptic) communication? Or did viruses co-opt retrotransposon gags that animals had previously co-opted for intercellular use?
academicjobsonline.org/ajo/jobs/25641
pop genomics, imaging, stat. genetics, machine learning aka cool science w big data.
academicjobsonline.org/ajo/jobs/25641
pop genomics, imaging, stat. genetics, machine learning aka cool science w big data.

http://academicjobsonline.org/ajo/jobs/25423
http://academicjobsonline.org/ajo/jobs/25423
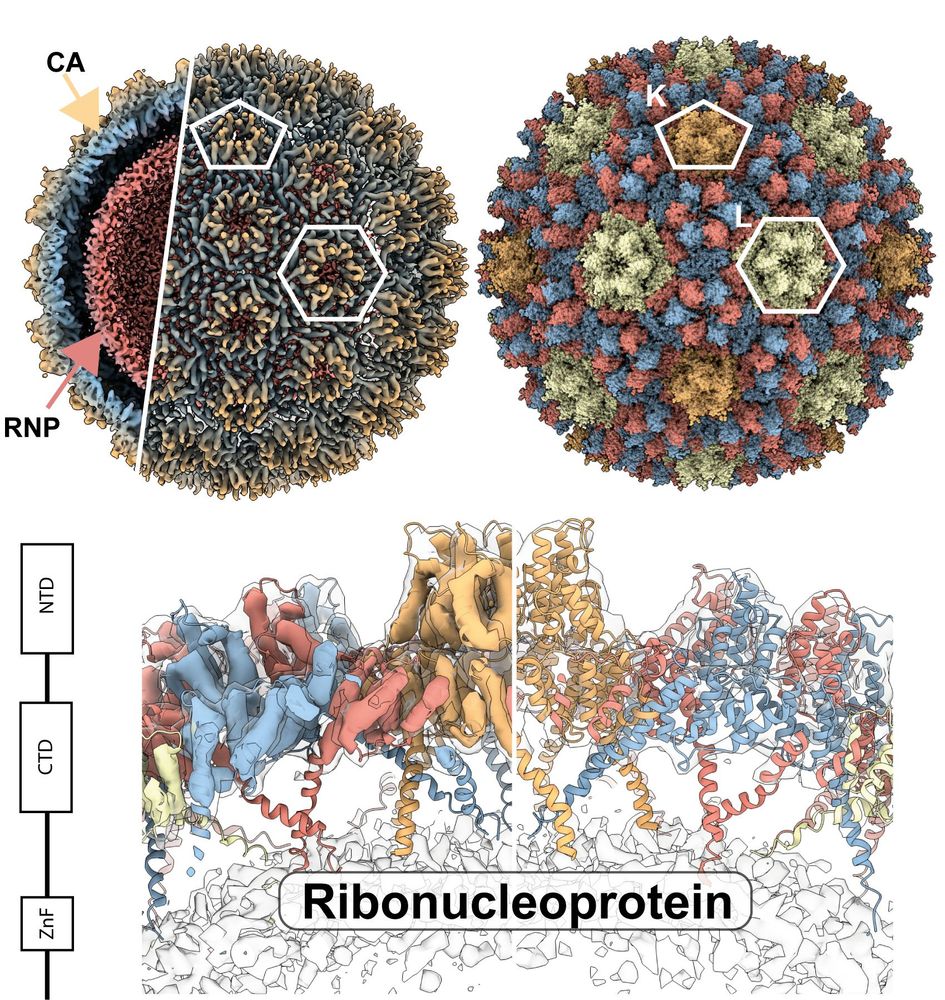
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

