PostDoc at the Jacobs-Wagner lab in Stanford. Previously at the Medalia Lab (Zurich) and Chlanda lab (Heidelberg). #teamtomo
www.nature.com/articles/s41...
And of course, all was only possible thanks to our great collaboration with @mizrahilab.bsky.social
(4/4)

www.nature.com/articles/s41...
And of course, all was only possible thanks to our great collaboration with @mizrahilab.bsky.social
(4/4)



Through the review process, we could refine our manuscript in several important areas, for example: (1/4)
Through the review process, we could refine our manuscript in several important areas, for example: (1/4)
It was a particular pleasure to work with a multi-disciplinary and international team on this project, from the University of Zurich, Ben-Gurion University in Beer-Sheva and the University of Greifswald!
It was a particular pleasure to work with a multi-disciplinary and international team on this project, from the University of Zurich, Ben-Gurion University in Beer-Sheva and the University of Greifswald!
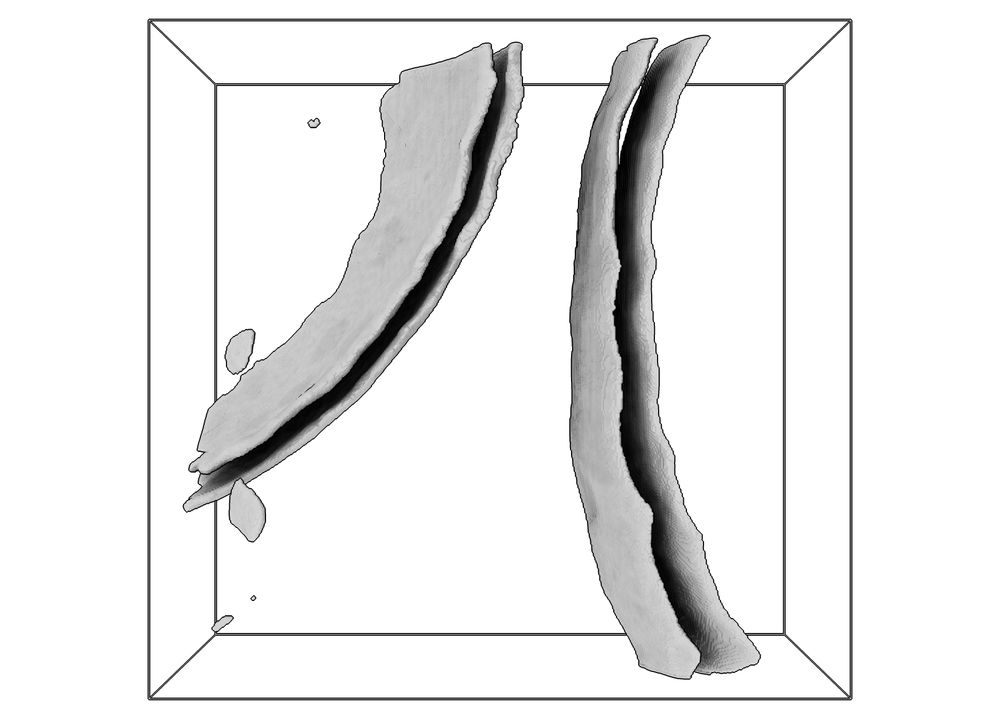
By combining in-situ #cryoet and #proteomics, we could assemble an integrative model, which illustrates the remarkable architecture of the amylosome - the complex that allows R. bromii to act as keystone degrader in the gut microbiome.
By combining in-situ #cryoet and #proteomics, we could assemble an integrative model, which illustrates the remarkable architecture of the amylosome - the complex that allows R. bromii to act as keystone degrader in the gut microbiome.
We wanted to understand, why it makes such a complicated assembly, and found through structural and biochemical analysis that two key enzymes act synergistically if they are bound in amylosome-like complexes. #proteomics revealed that this complex is enriched when needed.
We wanted to understand, why it makes such a complicated assembly, and found through structural and biochemical analysis that two key enzymes act synergistically if they are bound in amylosome-like complexes. #proteomics revealed that this complex is enriched when needed.
Our gut microbiome is essential for our health, and it relies on dietary fiber to thrive. The nutrients in dietary fiber, however, are very hard to access. Only few bacterial species manage to do so!
Our subject, R. bromii, is one of them - by using an extracellular assembly called amylosome.
Our gut microbiome is essential for our health, and it relies on dietary fiber to thrive. The nutrients in dietary fiber, however, are very hard to access. Only few bacterial species manage to do so!
Our subject, R. bromii, is one of them - by using an extracellular assembly called amylosome.
A short explainer below (1/5).
www.biorxiv.org/content/10.1...
#microbiology #microbiome #teamtomo
A short explainer below (1/5).
www.biorxiv.org/content/10.1...
#microbiology #microbiome #teamtomo
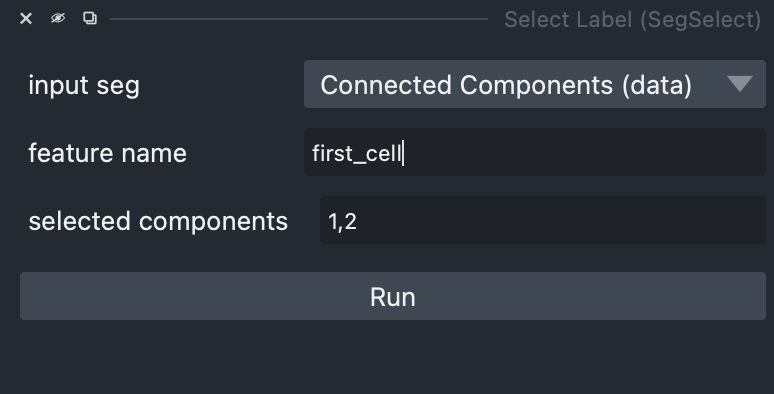
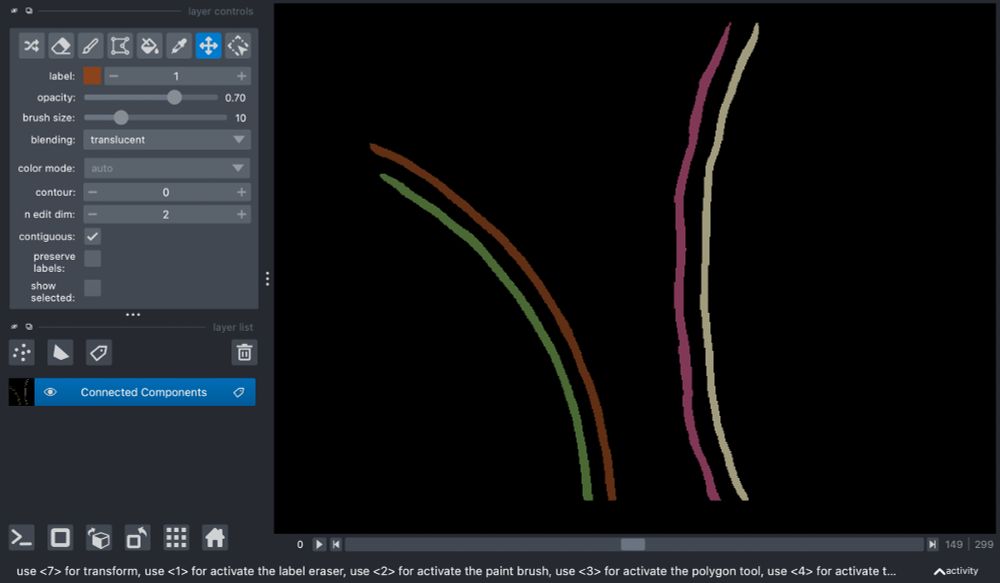
and the GitHub repository here: github.com/bwmr/napari-...
and the GitHub repository here: github.com/bwmr/napari-...