PostDoc at the Jacobs-Wagner lab in Stanford. Previously at the Medalia Lab (Zurich) and Chlanda lab (Heidelberg). #teamtomo



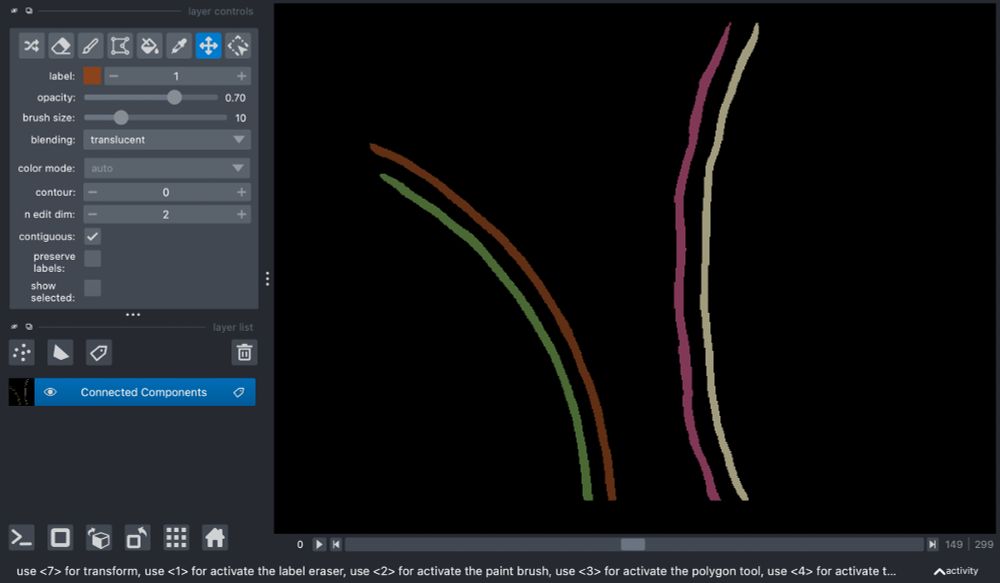
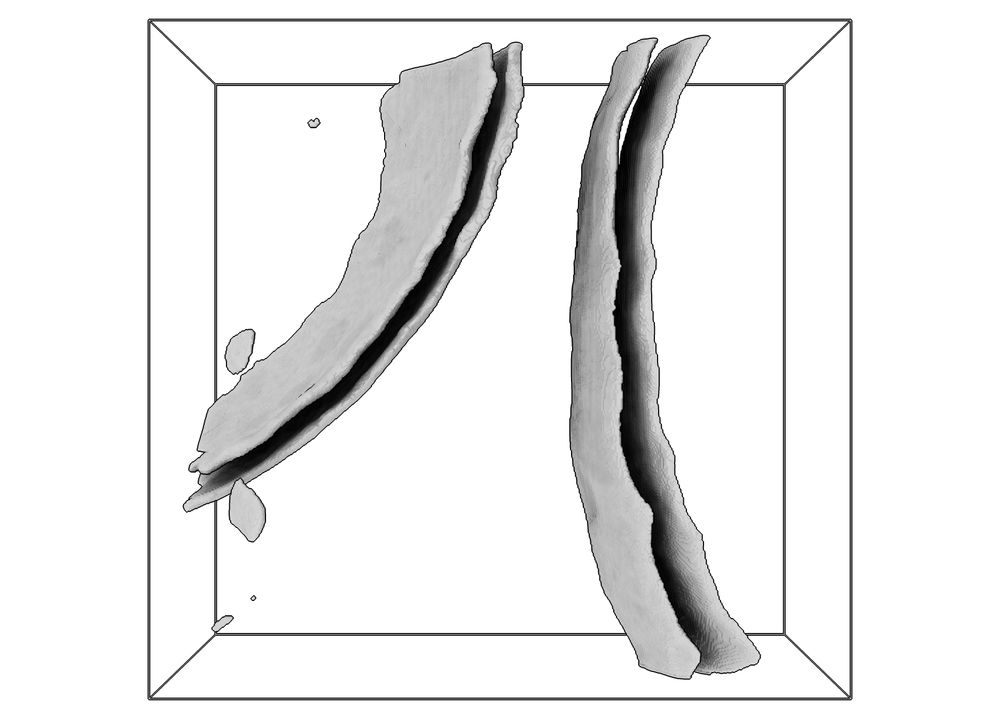
By combining in-situ #cryoet and #proteomics, we could assemble an integrative model, which illustrates the remarkable architecture of the amylosome - the complex that allows R. bromii to act as keystone degrader in the gut microbiome.
By combining in-situ #cryoet and #proteomics, we could assemble an integrative model, which illustrates the remarkable architecture of the amylosome - the complex that allows R. bromii to act as keystone degrader in the gut microbiome.