Former CIHMID Postdoc Fellow at Cornell. Stanford M&I Alum. NMDC Microbiome Data Champion @microbiomedata.org



Prehistoric Global Migration of Vanishing Gut Microbes With Humans
www.biorxiv.org/content/10.1...

Prehistoric Global Migration of Vanishing Gut Microbes With Humans
www.biorxiv.org/content/10.1...
recruit.ap.uci.edu/JPF09601

recruit.ap.uci.edu/JPF09601
May 10–12, 2026 | Bruges, Belgium
by @cp-immunity.bsky.social @cp-cellhostmicrobe.bsky.social and @cp-trendsmicrobiol.bsky.social
www.cell-symposia.com/microbiome-2...

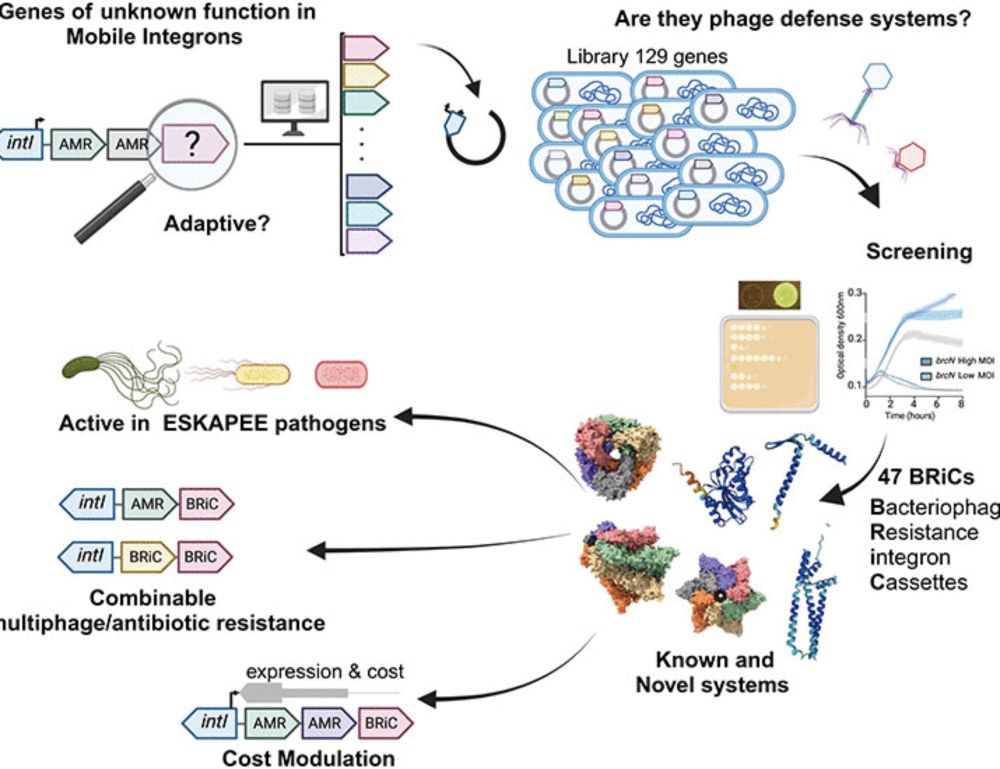
Microbial integrons in focus 🦠🧬
Two studies showing microbial integrons encoding phage defense systems
#MicroSky
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
Microbial integrons in focus 🦠🧬
Two studies showing microbial integrons encoding phage defense systems
#MicroSky
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
Has anyone developed a lab policy for for AI use in their biomedical research that they'd be willing to share?
Have people even thought about defining what should be allowed/encouraged, and what should be outright banned?
Please share with anyone who might have ideas.
Has anyone developed a lab policy for for AI use in their biomedical research that they'd be willing to share?
Have people even thought about defining what should be allowed/encouraged, and what should be outright banned?
Please share with anyone who might have ideas.
We call for a shift from disease surveillance to microbial stewardship, and highlight testable, cross-system hypotheses to unravel central rules of microbial life–spanning multiple scales, taxa, and environments. 🦠🌎
journals.asm.org/doi/10.1128/...

We call for a shift from disease surveillance to microbial stewardship, and highlight testable, cross-system hypotheses to unravel central rules of microbial life–spanning multiple scales, taxa, and environments. 🦠🌎
journals.asm.org/doi/10.1128/...
“We are deeply concerned about the current funding climate. When we think about new professors or postdocs starting their own labs, we worry that funding challenges could easily result in losing a generation of talented scientists.”
news.stanford.edu/stories/2025...

“We are deeply concerned about the current funding climate. When we think about new professors or postdocs starting their own labs, we worry that funding challenges could easily result in losing a generation of talented scientists.”
news.stanford.edu/stories/2025...
How many cells transmit when a strain is shared?
Can strain composition be dynamic when species composition is stable?
We answer these and related questions for the facial skin microbiome in our latest paper.
🧵[1/10]

How many cells transmit when a strain is shared?
Can strain composition be dynamic when species composition is stable?
We answer these and related questions for the facial skin microbiome in our latest paper.
🧵[1/10]
Wake Forest University School of Medicine affiliates can access @statnews.com for the next 2 weeks!
Visit www.statnews.com/register/ & enter a valid @wakehealth.edu email address
@wakeforest.bsky.social

Wake Forest University School of Medicine affiliates can access @statnews.com for the next 2 weeks!
Visit www.statnews.com/register/ & enter a valid @wakehealth.edu email address
@wakeforest.bsky.social

Check out this awesome video on the social #microbiome.
asm.org/Videos/Are-M...
@asm.org
Check out this awesome video on the social #microbiome.
asm.org/Videos/Are-M...
@asm.org
www.biorxiv.org/content/10.1...

The Sprockett Lab will focus on understanding the assembly, transmission, and evolution of the microbiome, and how these forces impact host health and physiology.
The Sprockett Lab will focus on understanding the assembly, transmission, and evolution of the microbiome, and how these forces impact host health and physiology.
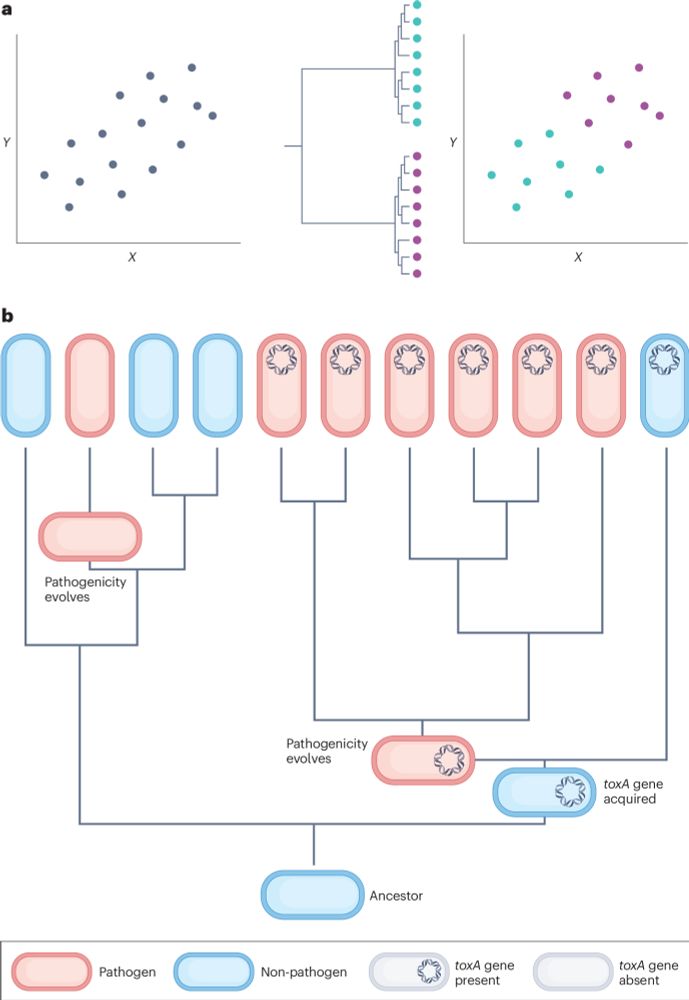
We show how phylogeny-based methods can resolve the problem of non-independence in genomic datasets.
These methods must be considered an essential part of the comparative genomics toolkit.
@lauriebelch.bsky.social @stuwest.bsky.social
We show how phylogeny-based methods can resolve the problem of non-independence in genomic datasets.
These methods must be considered an essential part of the comparative genomics toolkit.
@lauriebelch.bsky.social @stuwest.bsky.social
"Metagenomic immunoglobulin sequencing reveals IgA coating of microbial strains in the healthy human gut"
www.nature.com/articles/s41...

"Metagenomic immunoglobulin sequencing reveals IgA coating of microbial strains in the healthy human gut"
www.nature.com/articles/s41...
Nov 6th, 2023
10AM – noon, Corson-Mudd Hall, Room A409
Topics Include:
- Standardized Workflows for Metagenomic Analysis
- Metadata Management
- F.A.I.R. Data Use
Pre-Register Here: tinyurl.com/cornellmicrobiome
#microsky 🦠 🧫
Nov 6th, 2023
10AM – noon, Corson-Mudd Hall, Room A409
Topics Include:
- Standardized Workflows for Metagenomic Analysis
- Metadata Management
- F.A.I.R. Data Use
Pre-Register Here: tinyurl.com/cornellmicrobiome
#microsky 🦠 🧫

