Build computational models to (help) solve biology? Join us! https://www.deboramarkslab.com
DM or mail me!
popEVE (pop.evemodel.org) finds the needles in the haystacks of human genetic variation:

popEVE (pop.evemodel.org) finds the needles in the haystacks of human genetic variation:
🌐 evedesign.bio
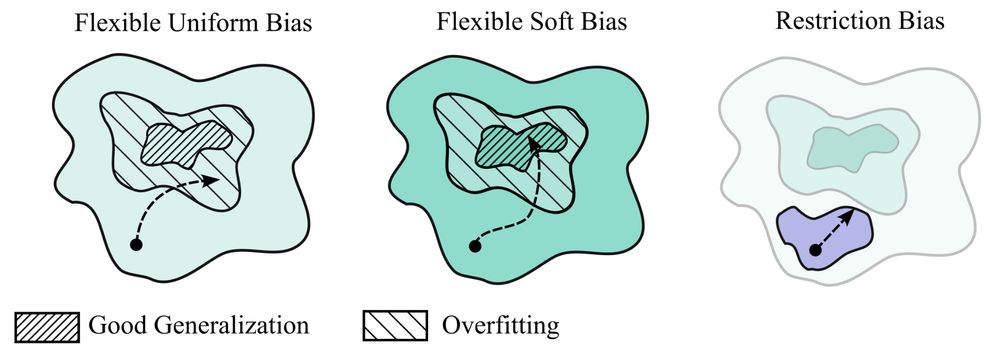
read all about it! biorxiv.org/content/10.110…
Been a long time goal of @deboramarks.bsky.social We tried a million years ago,
sciencedirect.com/science/articl���
Now total refresh led by Rohit Arora and Murphy Angelo and Pascal Notin for pulling together!!
read all about it! biorxiv.org/content/10.110…
Been a long time goal of @deboramarks.bsky.social We tried a million years ago,
sciencedirect.com/science/articl���
Now total refresh led by Rohit Arora and Murphy Angelo and Pascal Notin for pulling together!!
Huge thx to team for pushing on this when - led by Rohit Arora and Murphy Angelo for seeing this thru and Pascal Notin for pulling together!!!
🧵 1/9
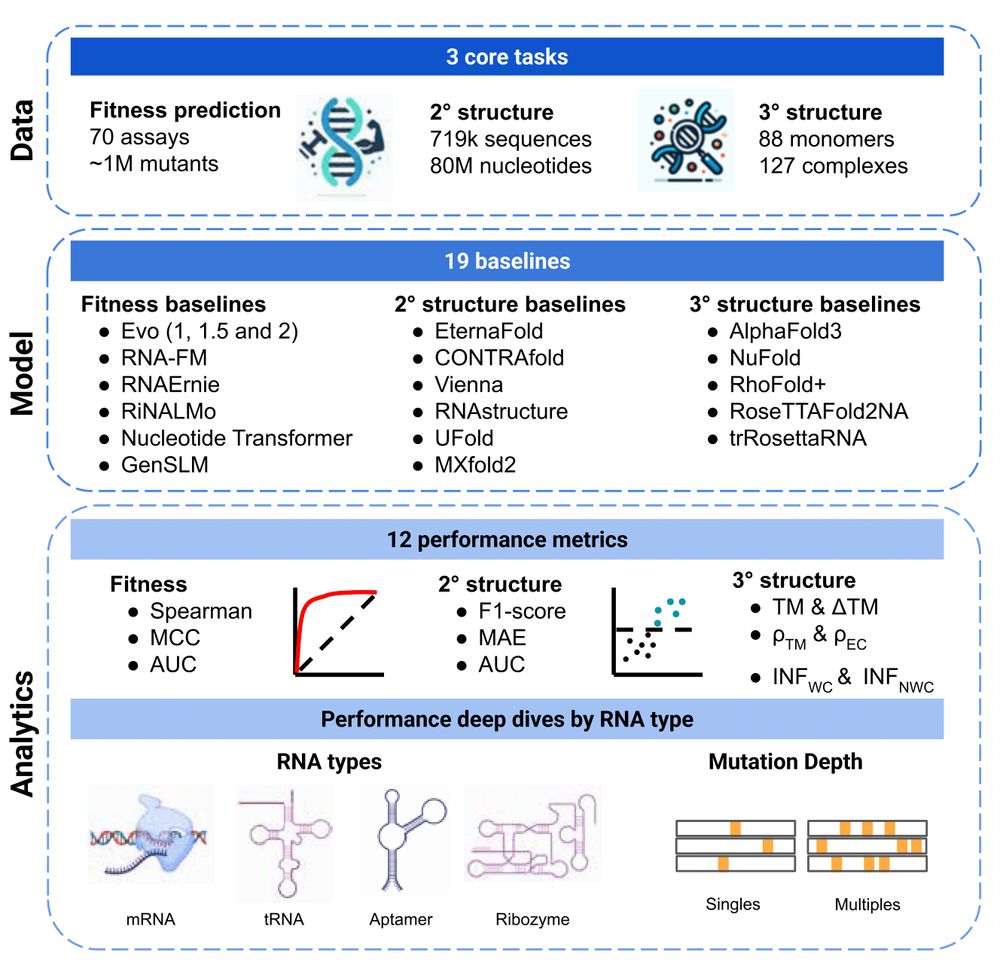
Huge thx to team for pushing on this when - led by Rohit Arora and Murphy Angelo for seeing this thru and Pascal Notin for pulling together!!!
@yaringal.bsky.social @deboramarks.bsky.social @pascalnotin.bsky.social
arxiv.org/abs/2506.089...
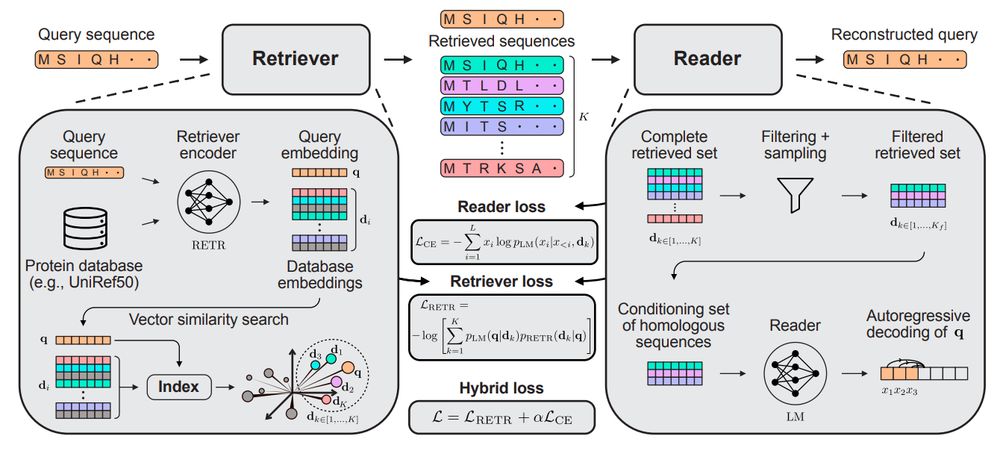
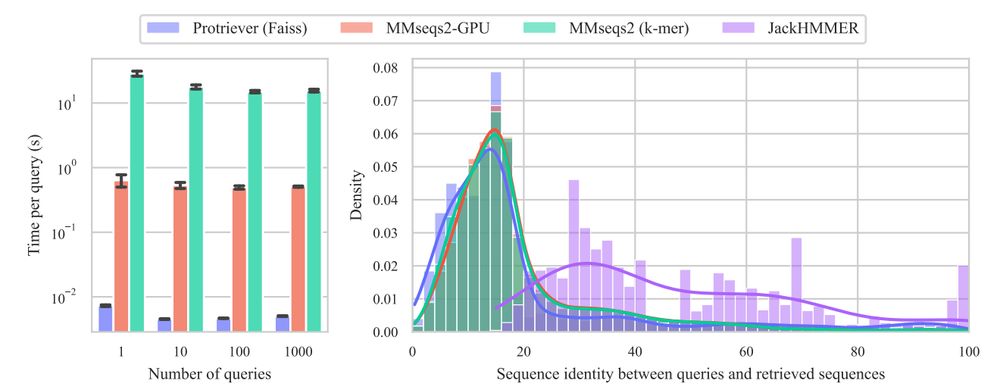
@yaringal.bsky.social @deboramarks.bsky.social @pascalnotin.bsky.social
arxiv.org/abs/2506.089...

fermatslibrary.com/journal_club

fermatslibrary.com/journal_club

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...




