

We built a proteome-wide model that combines cross-species and human population variation to rank missense variants by disease severity and help diagnose rare genetic disorders.
rdcu.be/eRu7K
Today, we introduce a simple algorithm that enables the model to learn from any rich feedback!
And then turns it into dense supervision.
(1/n)

Today, we introduce a simple algorithm that enables the model to learn from any rich feedback!
And then turns it into dense supervision.
(1/n)
Three papers on self-distillation, all from the same week!
arxiv.org/abs/2601.18734
arxiv.org/abs/2601.19897
arxiv.org/abs/2601.20802

Three papers on self-distillation, all from the same week!
arxiv.org/abs/2601.18734
arxiv.org/abs/2601.19897
arxiv.org/abs/2601.20802
Sharing our new paper @iclr_conf led by Yedi Zhang with Peter Latham
arxiv.org/abs/2512.20607
Sharing our new paper @iclr_conf led by Yedi Zhang with Peter Latham
arxiv.org/abs/2512.20607
arxiv.org/abs/2601.22950

arxiv.org/abs/2601.22950
www.pnas.org/doi/10.1073/...
www.pnas.org/doi/10.1073/...


Co-led by Noor Youssef and me, along with co-authors Navami Jain, Aarushi Mehrotra, Sarrah Leung, Abigail Jackson, @deboramarks.bsky.social, and with @cepi.net @futurehousesf.bsky.social!
Can protein language models help us fight viral outbreaks? Not yet. Here’s why 🧵👇
1/12

Co-led by Noor Youssef and me, along with co-authors Navami Jain, Aarushi Mehrotra, Sarrah Leung, Abigail Jackson, @deboramarks.bsky.social, and with @cepi.net @futurehousesf.bsky.social!
We're looking for a research tech to work on alt splicing, pancreatic islets and diabetes. The goal is to set a high-throughput platform to investigate the role of alternative exons in beta cell biology!
Interested in joining our lab at @melisupf.bsky.social? 👇
www.upf.edu/documents/d/...
We're looking for a research tech to work on alt splicing, pancreatic islets and diabetes. The goal is to set a high-throughput platform to investigate the role of alternative exons in beta cell biology!
Interested in joining our lab at @melisupf.bsky.social? 👇
www.upf.edu/documents/d/...
A thread... 🧵
www.medrxiv.org/content/10.1...

A thread... 🧵
www.medrxiv.org/content/10.1...
Mafalda, @cwjpugh.bsky.social and I will be in San Diego next week for NeurIPS -- happy to chat variant effect prediction (or just say hi).
“From Likelihood to Fitness: Improving Variant Effect Prediction in Protein and Genome Language Models”
openreview.net/pdf/a151f62e...
Mafalda, @cwjpugh.bsky.social and I will be in San Diego next week for NeurIPS -- happy to chat variant effect prediction (or just say hi).
“From Likelihood to Fitness: Improving Variant Effect Prediction in Protein and Genome Language Models”
openreview.net/pdf/a151f62e...
popEVE (pop.evemodel.org) finds the needles in the haystacks of human genetic variation:

popEVE (pop.evemodel.org) finds the needles in the haystacks of human genetic variation:
We built a proteome-wide model that combines cross-species and human population variation to rank missense variants by disease severity and help diagnose rare genetic disorders.
rdcu.be/eRu7K

We built a proteome-wide model that combines cross-species and human population variation to rank missense variants by disease severity and help diagnose rare genetic disorders.
rdcu.be/eRu7K

Learn more about the PhD programme and how to apply → www.evomg-dn.eu
#PhD #DoctoralTraining #ResearchCareers #LifeSciences #Genomics #EvolutionaryBiology

Learn more about the PhD programme and how to apply → www.evomg-dn.eu
#PhD #DoctoralTraining #ResearchCareers #LifeSciences #Genomics #EvolutionaryBiology
Join us to develop deep generative models of cross-species data to tackle open questions in disease genetics.
www.crg.eu/en/content/t...
Be part of EvoMG-DN and advance your PhD career in evolutionary genomics.
👉 www.evomg-dn.eu
#PhD #DoctoralTraining #ResearchCareers #LifeSciences #BiomedicalResearch #Genomics #EvolutionaryBiology #MSCA #HorizonEurope #EUResearch #ResearchOpportunities #PhDPositions

Be part of EvoMG-DN and advance your PhD career in evolutionary genomics.
👉 www.evomg-dn.eu
#PhD #DoctoralTraining #ResearchCareers #LifeSciences #BiomedicalResearch #Genomics #EvolutionaryBiology #MSCA #HorizonEurope #EUResearch #ResearchOpportunities #PhDPositions
We move beyond reconstruction to learn rich, cell-level embeddings for diverse omics.
📘 www.biorxiv.org/content/10.1...

We move beyond reconstruction to learn rich, cell-level embeddings for diverse omics.
📘 www.biorxiv.org/content/10.1...
I am excited to announce that our new study explaining the missing heritability of many phenotypes using WGS data from ~347,000 UK Biobank participants has just been published in @Nature.
Our manuscript is here: www.nature.com/articles/s41....

I am excited to announce that our new study explaining the missing heritability of many phenotypes using WGS data from ~347,000 UK Biobank participants has just been published in @Nature.
Our manuscript is here: www.nature.com/articles/s41....


We show how phylogeny-based methods can resolve the problem of non-independence in genomic datasets.
These methods must be considered an essential part of the comparative genomics toolkit.
@lauriebelch.bsky.social @stuwest.bsky.social
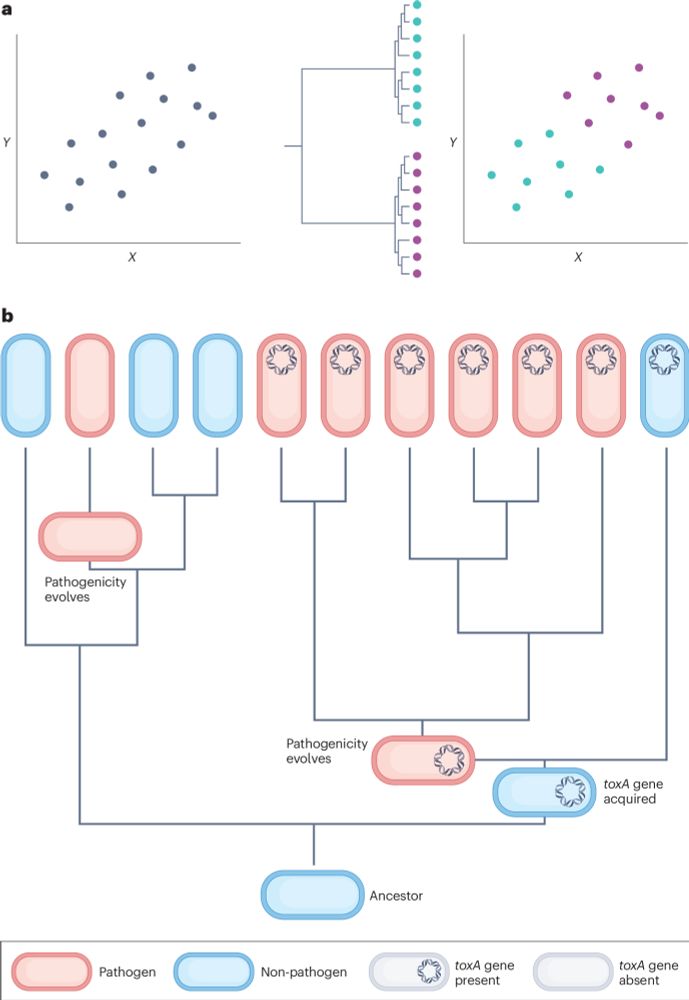
We show how phylogeny-based methods can resolve the problem of non-independence in genomic datasets.
These methods must be considered an essential part of the comparative genomics toolkit.
@lauriebelch.bsky.social @stuwest.bsky.social
Want to change the consequences of receptor activation?
Small molecules binding the GPCR-transducer interface change G protein subtype preference in predictable ways, enabling rational drug design 💥
So many new possibilities! 🧪🧠🟦
www.nature.com/articles/s41...
🧵👇

Want to change the consequences of receptor activation?
Small molecules binding the GPCR-transducer interface change G protein subtype preference in predictable ways, enabling rational drug design 💥
So many new possibilities! 🧪🧠🟦
www.nature.com/articles/s41...
🧵👇
Join us to develop deep generative models of cross-species data to tackle open questions in disease genetics.
www.crg.eu/en/content/t...

Join us to develop deep generative models of cross-species data to tackle open questions in disease genetics.
www.crg.eu/en/content/t...
• End-to-end protein design for everyone!
• Analyze your generated library interactively and on 3D structures
• Export codon-optimized DNA sequences for experimental testing.
• End-to-end protein design for everyone!
• Analyze your generated library interactively and on 3D structures
• Export codon-optimized DNA sequences for experimental testing.

