
Visit hub.dsmz.de

You will be responsible for guiding the development of this important resource and for curating the SILVA taxonomy🌳and more!
👉 www.dsmz.de/dsmz/career/...
Read and share ‼️
#sciencejobs 🦠🧪 #career #taxonomy

You will be responsible for guiding the development of this important resource and for curating the SILVA taxonomy🌳and more!
👉 www.dsmz.de/dsmz/career/...
Read and share ‼️
#sciencejobs 🦠🧪 #career #taxonomy
📑The new publication about the #SILVA database for the #NAR database issue is now online.
👉 academic.oup.com/nar/article/...

📑The new publication about the #SILVA database for the #NAR database issue is now online.
👉 academic.oup.com/nar/article/...
The #SILVA team will inform about the upcoming release, the new website design 🖥️, and more!
Write to [email protected] to receive the Zoom login details
#science #FAIRdata 🧫🧪

The #SILVA team will inform about the upcoming release, the new website design 🖥️, and more!
Write to [email protected] to receive the Zoom login details
#science #FAIRdata 🧫🧪

We are especially looking forward to the opportunity to answer your questions, and hear your feedback and ideas. 🦠🧬.
Write to [email protected] to receive the Zoom login details 🖥️.

We are especially looking forward to the opportunity to answer your questions, and hear your feedback and ideas. 🦠🧬.
Write to [email protected] to receive the Zoom login details 🖥️.
academic.oup.com/nar/advance-...
@dsmzd3.bsky.social @leibniz-dsmz.bsky.social
#TYGS and #LPSN in 2025: a Global Core Biodata Resource 🧬🖥️ for genome-based classification and nomenclature of prokaryotes 🦠 within #DSMZDigitalDiversity
👉 academic.oup.com/nar/article/...

#TYGS and #LPSN in 2025: a Global Core Biodata Resource 🧬🖥️ for genome-based classification and nomenclature of prokaryotes 🦠 within #DSMZDigitalDiversity
👉 academic.oup.com/nar/article/...
Follow the link to read the report.
🔗https://www.microbiologyresearch.org/content/journal/ijsem/10.1099/ijsem.0.006943

Follow the link to read the report.
🔗https://www.microbiologyresearch.org/content/journal/ijsem/10.1099/ijsem.0.006943

🙏 Huge thanks to all participants and our community for 3 days of knowledge-sharing and collaboration.




🙏 Huge thanks to all participants and our community for 3 days of knowledge-sharing and collaboration.
Highlights included:
🎤 A thought-provoking keynote by Alexandre Smirnov
💬 A lively panel discussion on bridging lab practices and scientific publishing
🤝 Engaging networking and community exchange session. Stay tuner for day 3!



Highlights included:
🎤 A thought-provoking keynote by Alexandre Smirnov
💬 A lively panel discussion on bridging lab practices and scientific publishing
🤝 Engaging networking and community exchange session. Stay tuner for day 3!
Read all about #StrainInfo - the central database for linked microbial strain identifiers 🧫🖥️
👉 academic.oup.com/database/art...
#DSMZDigitalDiversity #science 🧪 #databases #FAIRdata @nfdi4microbiota.bsky.social 🧬

Read all about #StrainInfo - the central database for linked microbial strain identifiers 🧫🖥️
👉 academic.oup.com/database/art...
#DSMZDigitalDiversity #science 🧪 #databases #FAIRdata @nfdi4microbiota.bsky.social 🧬
#SILVA 🧬 joined @leibniz-dsmz.bsky.social 🧫 and the @dsmzd3.bsky.social in 2023. This is now also visible on the redesigned website 🖌️, which has been restructured reorganized in a first step towards an improved and modernized #SILVA!
Try it out at www.arb-silva.de

#SILVA 🧬 joined @leibniz-dsmz.bsky.social 🧫 and the @dsmzd3.bsky.social in 2023. This is now also visible on the redesigned website 🖌️, which has been restructured reorganized in a first step towards an improved and modernized #SILVA!
Try it out at www.arb-silva.de
A first prototype of a knowledge graph has been released and is now accessible through a newly established SPARQL endpoint: lnkd.in/eg_ZfTMY
The graph is based on the @dsmzd3.bsky.social Ontology and currently supports integration efforts within the @dsmzd3.bsky.social Hub.

A first prototype of a knowledge graph has been released and is now accessible through a newly established SPARQL endpoint: lnkd.in/eg_ZfTMY
The graph is based on the @dsmzd3.bsky.social Ontology and currently supports integration efforts within the @dsmzd3.bsky.social Hub.
@dsmzd3.bsky.social

@dsmzd3.bsky.social

Join us for updates and the opportunity to ask questions and provide feedback 🦠🧬.
Write to [email protected] to receive the Zoom login details 🖥️.

Join us for updates and the opportunity to ask questions and provide feedback 🦠🧬.
Write to [email protected] to receive the Zoom login details 🖥️.


🗣️ "From microbes to molecules: How DSMZ digital diversity integrates diverse datasets into a unified framework"
#ECCB2025 #Bioinformatics

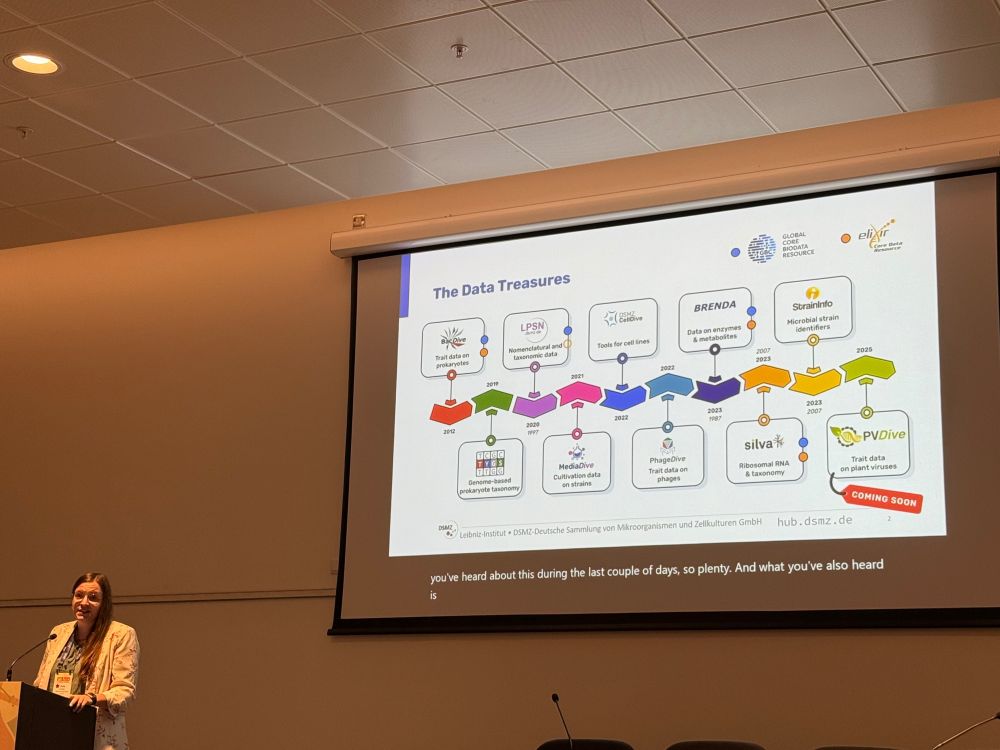
🗣️ "From microbes to molecules: How DSMZ digital diversity integrates diverse datasets into a unified framework"
#ECCB2025 #Bioinformatics


Isabel, Melania and Manuela are happy to answer your questions!
#FEMS2025 #DSMZDigitalDiversity #biodiversity @dsmzd3.bsky.social


Isabel, Melania and Manuela are happy to answer your questions!
#FEMS2025 #DSMZDigitalDiversity #biodiversity @dsmzd3.bsky.social
Join the 4th NFDI4Microbiota Conference in Cologne
📅 Sept 30–Oct 2 | Theme: From Lab to Publication
Hands-on workshops, keynotes, posters | Registration is free
📝 Register by Aug 31: events.hifis.net/eve...
#FAIRdata #Microbiome

Join the 4th NFDI4Microbiota Conference in Cologne
📅 Sept 30–Oct 2 | Theme: From Lab to Publication
Hands-on workshops, keynotes, posters | Registration is free
📝 Register by Aug 31: events.hifis.net/eve...
#FAIRdata #Microbiome

