📄 doi.org/10.1038/s415...
👤 EVBC: Janusz Bujnicki

📄 doi.org/10.1038/s415...
👤 EVBC: Janusz Bujnicki

ClaPNAC identifies various types of nucleotide-aminoacid contacts in 3D structures of NA-protein complexes.
More in the preprint: doi.org/10.1101/2025...
#Bioinformatics #Protein #RNA #DNA #Structure #Contact #3D

ClaPNAC identifies various types of nucleotide-aminoacid contacts in 3D structures of NA-protein complexes.
More in the preprint: doi.org/10.1101/2025...
#Bioinformatics #Protein #RNA #DNA #Structure #Contact #3D
#MedSky #STS

doi.org/10.1186/s130...
ARTEM enables screening RNA and DNA 3D structure databases for arbitrary 3D motifs. Kink-turns, G-quadruplexes, GNRA tetraloops, i-motifs, you name it!
#Research #Bioinformatics #3D #RNA #DNA

doi.org/10.1186/s130...
ARTEM enables screening RNA and DNA 3D structure databases for arbitrary 3D motifs. Kink-turns, G-quadruplexes, GNRA tetraloops, i-motifs, you name it!
#Research #Bioinformatics #3D #RNA #DNA
Read the preprint:
biorxiv.org/content/10.1...
#RNA #Structure #3D #GNRA #Tetraloop #Motif #Research #RNAprotein #Interactions

Read the preprint:
biorxiv.org/content/10.1...
#RNA #Structure #3D #GNRA #Tetraloop #Motif #Research #RNAprotein #Interactions
The Lab of #RNA #Algorithms @ imol.institute
has an open position for a #PostDoc interested in RNA computational biology.
Apply before July 10th: euraxess.ec.europa.eu/jobs/350755
Please spread the word!
#Academia #Position #Research #Bioinformatics #Poland #Warsaw #Hiring
The Lab of #RNA #Algorithms @ imol.institute
has an open position for a #PostDoc interested in RNA computational biology.
Apply before July 10th: euraxess.ec.europa.eu/jobs/350755
Please spread the word!
#Academia #Position #Research #Bioinformatics #Poland #Warsaw #Hiring
Looking to tackle computational challenges of RNA folding and binding? Come build new RNA algorithms as a #postdoc in my lab!
More info: euraxess.ec.europa.eu/jobs/350755
#Academia #Position #Job #Research #RNA #Structure #3D #Bioinformatics #Poland #Warsaw #IMolPAS #NCN

Looking to tackle computational challenges of RNA folding and binding? Come build new RNA algorithms as a #postdoc in my lab!
More info: euraxess.ec.europa.eu/jobs/350755
#Academia #Position #Job #Research #RNA #Structure #3D #Bioinformatics #Poland #Warsaw #IMolPAS #NCN
Details here: euraxess.ec.europa.eu/jobs/350755
#postdoc #Academia #Position #Job #Research #RNA #Structure #Bioinformatics #Poland #Warsaw #IMolPAS #NCN

Details here: euraxess.ec.europa.eu/jobs/350755
#postdoc #Academia #Position #Job #Research #RNA #Structure #Bioinformatics #Poland #Warsaw #IMolPAS #NCN
github.com/febos/SQUARNA
Updated preprint is coming soon :)
#SQUARNA #RNA #2D #Structure #Prediction #Bioinformatics #Research #Pseudoknot
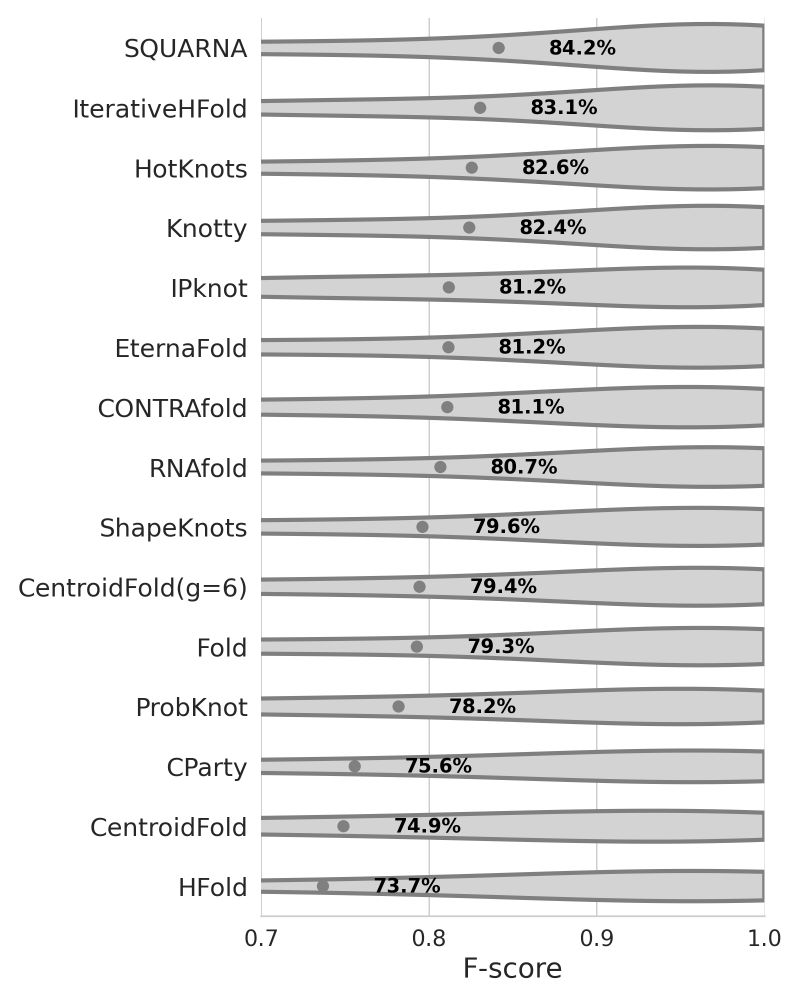
github.com/febos/SQUARNA
Updated preprint is coming soon :)
#SQUARNA #RNA #2D #Structure #Prediction #Bioinformatics #Research #Pseudoknot
More weird kink-turns in our recent preprint:
doi.org/10.1101/2024...
#RNA #ARTEM #Kinkturn #Kturn #Motif #Bioinformatics #Science #Research #StructuralBiology

More weird kink-turns in our recent preprint:
doi.org/10.1101/2024...
#RNA #ARTEM #Kinkturn #Kturn #Motif #Bioinformatics #Science #Research #StructuralBiology
Extended deadline: March 24, 2025.

Extended deadline: March 24, 2025.
github.com/david-bogdan...
#RNA #DNA #structure #3d #alignment #bioinformatics #research #PDB #tutorial
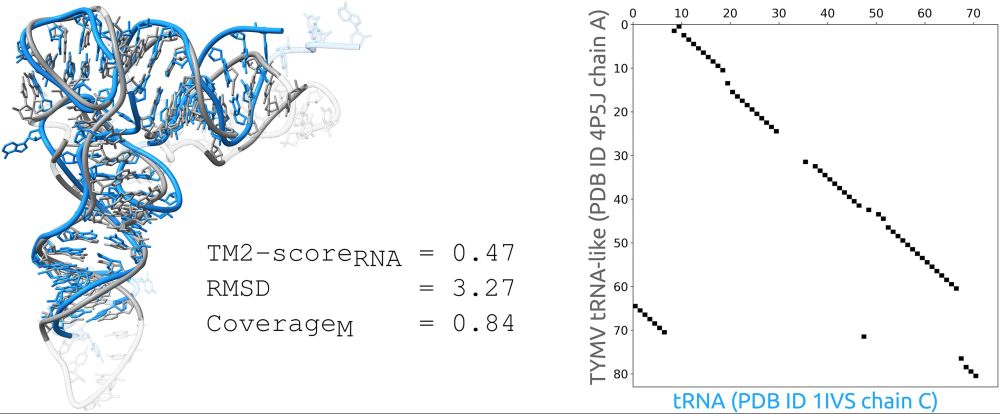
github.com/david-bogdan...
#RNA #DNA #structure #3d #alignment #bioinformatics #research #PDB #tutorial
Meet MIXALIME, a framework for assessing allelic imbalance, and UDACHA, a database of allele-specific chromatin accessibility, read more at www.nature.com/articles/s41...

doi.org/10.1093/nar/...
DesiRNA is a tool for RNA sequence design (inverse folding) and it is the first to solve all 100 puzzles in the Eterna100 benchmark within 24 h!
#RNA #Design #Research #Bioinformatics #Science #Structure

doi.org/10.1093/nar/...
DesiRNA is a tool for RNA sequence design (inverse folding) and it is the first to solve all 100 puzzles in the Eterna100 benchmark within 24 h!
#RNA #Design #Research #Bioinformatics #Science #Structure
Find out more:
doi.org/10.1093/nar/...
#research

Find out more:
doi.org/10.1093/nar/...
#research
Find out more:
doi.org/10.1093/nar/...
#RNA #research #Bioinformatics #motif #Staple

Find out more:
doi.org/10.1093/nar/...
#RNA #research #Bioinformatics #motif #Staple
www.nature.com/articles/s41...
It reports the evaluation of blind prediction of 23 targets predicted by 18 groups worldwide. I'm glad that our group ranked as one of the top four groups in this competition!
#RNA #prediction #RNApuzzles #CASP16

www.nature.com/articles/s41...
It reports the evaluation of blind prediction of 23 targets predicted by 18 groups worldwide. I'm glad that our group ranked as one of the top four groups in this competition!
#RNA #prediction #RNApuzzles #CASP16
Find out more:
doi.org/10.1016/j.jm...
#RNA #research #Structure #cryoEM #strand #motif #raiA

Find out more:
doi.org/10.1016/j.jm...
#RNA #research #Structure #cryoEM #strand #motif #raiA
pip install SQUARNA
import SQUARNA
SQUARNA.Predict(inputseq="CCCGNRAGGG")
Demo:
github.com/febos/SQUARN...
BioRxiv:
doi.org/10.1101/2023...
#RNA #Structure #Bioinformatics #Research #Prediction #2D #pip #Python

pip install SQUARNA
import SQUARNA
SQUARNA.Predict(inputseq="CCCGNRAGGG")
Demo:
github.com/febos/SQUARN...
BioRxiv:
doi.org/10.1101/2023...
#RNA #Structure #Bioinformatics #Research #Prediction #2D #pip #Python
Find out which is which in our recent preprint on RNA tertiary motif search and their isostericity:
doi.org/10.1101/2024...
#rna #dna #3d

Find out which is which in our recent preprint on RNA tertiary motif search and their isostericity:
doi.org/10.1101/2024...
#rna #dna #3d
ARTEMIS is a tool for #RNA/#DNA #3D #structure #superposition and structure-based #sequence #alignment.
It allowed us to identify an intriguing structural similarity between Lysine and M-box riboswitches!
doi.org/10.1093/nar/...
#Bioinformatics

ARTEMIS is a tool for #RNA/#DNA #3D #structure #superposition and structure-based #sequence #alignment.
It allowed us to identify an intriguing structural similarity between Lysine and M-box riboswitches!
doi.org/10.1093/nar/...
#Bioinformatics
🚀Check out kink-turn-like motifs with no kink, and crazy kink-turn variants in group II introns.🔬
doi.org/10.1101/2024...
github.com/febos/ARTEM-KT
github.com/david-bogdan...
#Science #Bioinformatics #Research #Structure

🚀Check out kink-turn-like motifs with no kink, and crazy kink-turn variants in group II introns.🔬
doi.org/10.1101/2024...
github.com/febos/ARTEM-KT
github.com/david-bogdan...
#Science #Bioinformatics #Research #Structure

