
http://g-evol.com
See
job-portal.lmu.de/jobposting/6...
See
job-portal.lmu.de/jobposting/6...
See
job-portal.lmu.de/jobposting/6...
👉 bornberglab.org/post-doc-pos...
👉 bornberglab.org/post-doc-pos...
doi.org/10.1093/jeb/...
Erwann et al. 2025

doi.org/10.1093/jeb/...
Erwann et al. 2025
🤝 You will be part of the DFG SPP 2349 @gevol.bsky.social consortium & the @imprsgs.bsky.social, with research visits to collaboration partners (👇).
🤝 You will be part of the DFG SPP 2349 @gevol.bsky.social consortium & the @imprsgs.bsky.social, with research visits to collaboration partners (👇).
Join us at @uni-goettingen.de for a 3-year PhD (65% TV-L E13).
We will investigate the genomic & phenotypic impact of gene duplication across 233 arthropod genomes!
More infos 👇 and s.gwdg.de/eDrAAY
#Evolution #Genomics #Bioinformatics
Join us at @uni-goettingen.de for a 3-year PhD (65% TV-L E13).
We will investigate the genomic & phenotypic impact of gene duplication across 233 arthropod genomes!
More infos 👇 and s.gwdg.de/eDrAAY
#Evolution #Genomics #Bioinformatics
We present a comprehensive review of de novo gene emergence — providing a classification of current detection methods and a roadmap for addressing major challenges in the field of gene birth from non-genic sequences.

We present a comprehensive review of de novo gene emergence — providing a classification of current detection methods and a roadmap for addressing major challenges in the field of gene birth from non-genic sequences.
We introduce a user-friendly toolkit that implements our novel DeNoFo file-format for standardised annotation of de novo gene detection workflows — enabling reproducible methodology descriptions and easier dataset comparison across studies.

We introduce a user-friendly toolkit that implements our novel DeNoFo file-format for standardised annotation of de novo gene detection workflows — enabling reproducible methodology descriptions and easier dataset comparison across studies.
academic.oup.com/bioinformati...
academic.oup.com/bioinformati...
onlinelibrary.wiley.com/toc/15525015... 🚨

onlinelibrary.wiley.com/toc/15525015... 🚨



Thanks to everyone who presented, especially our invited speakers @ahuylmans.bsky.social & @rmwaterhouse.bsky.social. From GEvol, Marie Lebherz, Elisa Israel and Barbara Feldmeyer gave talks about their work. (1/2)




Thanks to everyone who presented, especially our invited speakers @ahuylmans.bsky.social & @rmwaterhouse.bsky.social. From GEvol, Marie Lebherz, Elisa Israel and Barbara Feldmeyer gave talks about their work. (1/2)



🔬 Ever wondered how new genes emerge from scratch? Meet DESwoMAN, a fully automated pipeline to detect and analyze newly expressed ORFs (neORFs) from transcriptome data — giving us a window into the earliest stages of de novo gene emergence!
(Thread 🧵👇)
🔬 Ever wondered how new genes emerge from scratch? Meet DESwoMAN, a fully automated pipeline to detect and analyze newly expressed ORFs (neORFs) from transcriptome data — giving us a window into the earliest stages of de novo gene emergence!
(Thread 🧵👇)
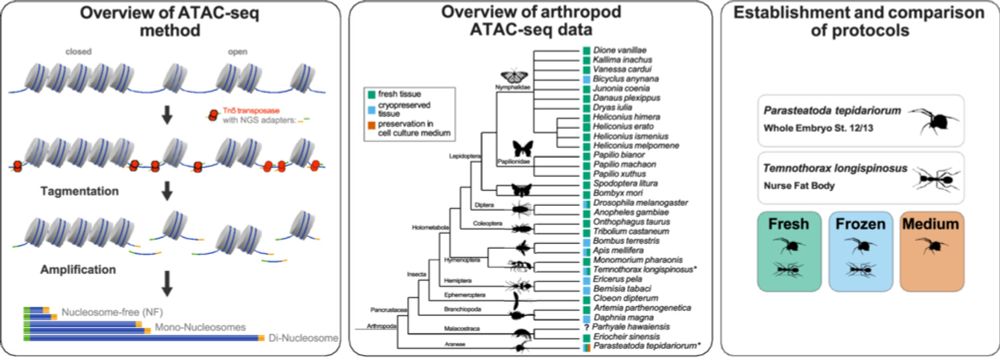
in JEZ-B.🕸️ doi.org/10.1002/jez....

in JEZ-B.🕸️ doi.org/10.1002/jez....

