@damiepak.bsky.social
Register or sponsor a student by donating to support Ukraine!
Details: bit.ly/3wBeY4S
Please share!
#AcademicSky #EconSky #RStats

@damiepak.bsky.social
Register or sponsor a student by donating to support Ukraine!
Details: bit.ly/3wBeY4S
Please share!
#AcademicSky #EconSky #RStats
Great for those who love pipelines, whole genome data and work with a social purpose.
ubc.wd10.myworkdayjobs.com/ubcfacultyjo...

Great for those who love pipelines, whole genome data and work with a social purpose.
ubc.wd10.myworkdayjobs.com/ubcfacultyjo...



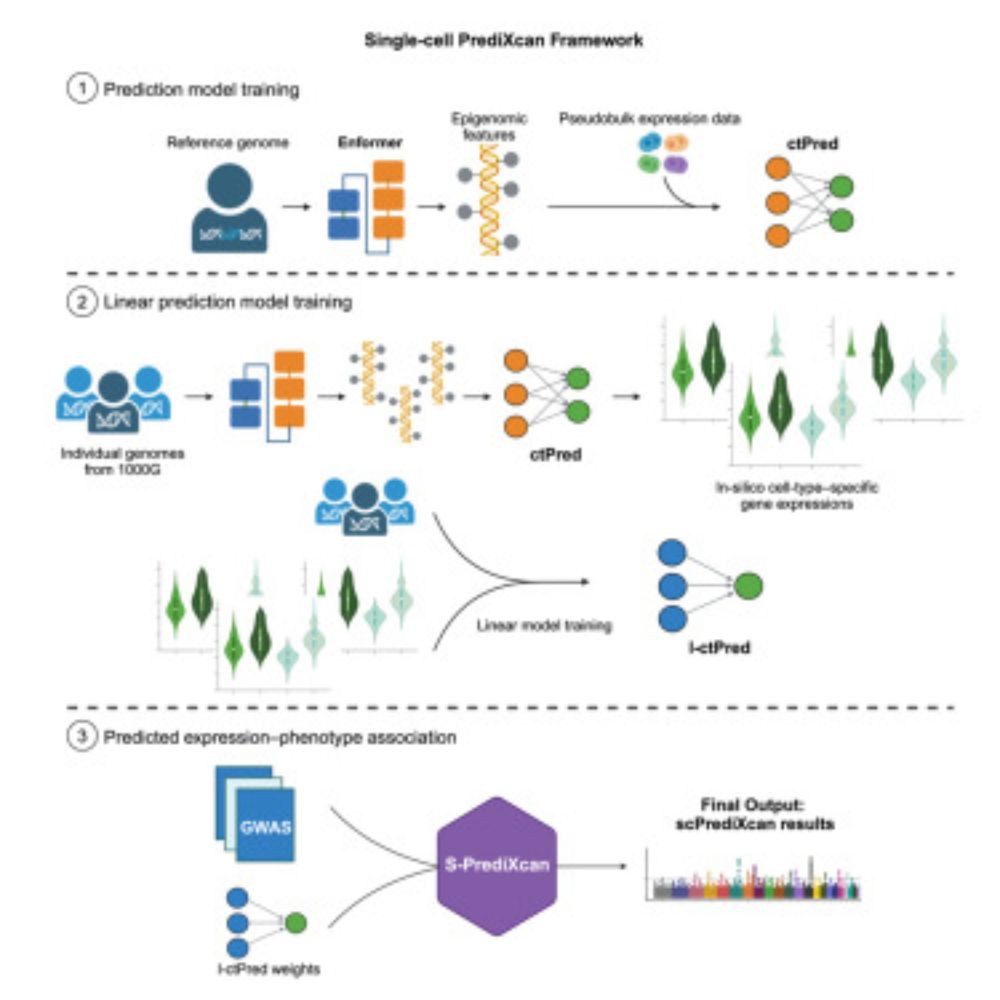
www.cell.com/cell-genomic...
Led by the talented @Charles_Zhou12 and supervised by @MengjieChen6
and me, with thanks to many contributors.
scPrediXcan integrates deep learning and single cell expression data into a powerful cell type specific TWAS framework.
www.cell.com/cell-genomic...
Led by the talented @Charles_Zhou12 and supervised by @MengjieChen6
and me, with thanks to many contributors.
scPrediXcan integrates deep learning and single cell expression data into a powerful cell type specific TWAS framework.
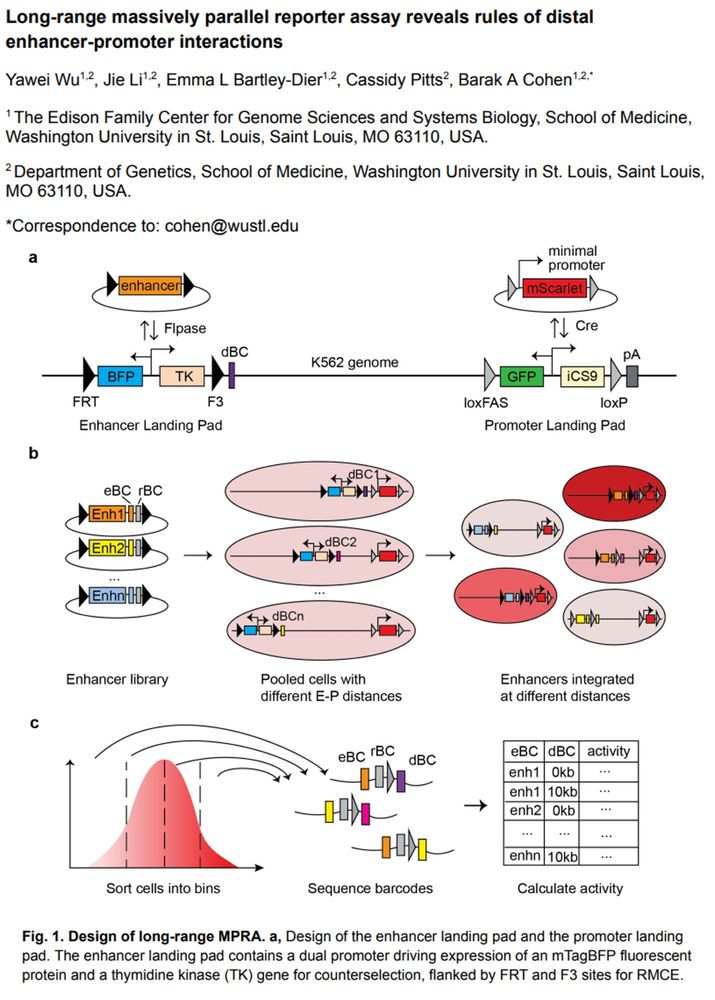
Long-range massively parallel reporter assay reveals rules of distal enhancer-promoter interactions
From Barak Cohen's lab at @washu.bsky.social
Read the pre-print 👇
doi.org/10.1101/2025...
Learn more about the research from the Cohen lab: bclab.wustl.edu
Currently cataloguing about 700k virus species!
📄 www.biorxiv.org/content/10.1...

Currently cataloguing about 700k virus species!
📄 www.biorxiv.org/content/10.1...
Rewriting regulatory DNA to dissect and reprogram gene expression
Our new method (Variant-EFFECTS) uses high-throughput prime editing + flow sorting + sequencing to precisely measure effects of noncoding variants on gene expression
Thread 👇

Rewriting regulatory DNA to dissect and reprogram gene expression
Our new method (Variant-EFFECTS) uses high-throughput prime editing + flow sorting + sequencing to precisely measure effects of noncoding variants on gene expression
Thread 👇
Shift augmentation improves DNA convolutional neural network indel effect predictions
www.biorxiv.org/content/10.1...

Shift augmentation improves DNA convolutional neural network indel effect predictions
www.biorxiv.org/content/10.1...

Research Associate / Staff Scientist
Vancouver Canada
Join the Silent Genomes Project to provide an outstanding genetic variation database for Indigenous peoples of Canada to improve rare disease diagnosis
tinyurl.com/bm4ptux3
Priority to Canadians
Please repost

Research Associate / Staff Scientist
Vancouver Canada
Join the Silent Genomes Project to provide an outstanding genetic variation database for Indigenous peoples of Canada to improve rare disease diagnosis
tinyurl.com/bm4ptux3
Priority to Canadians
Please repost
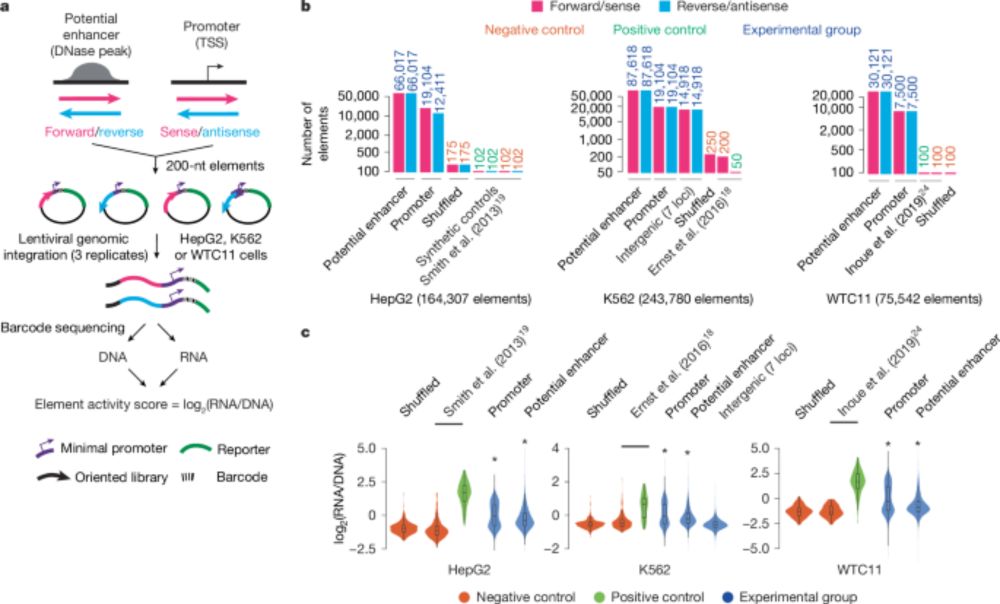
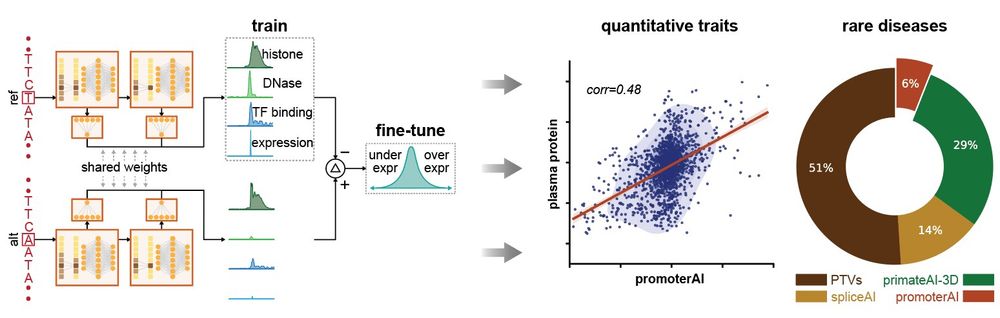
www.nature.com/articles/s41...
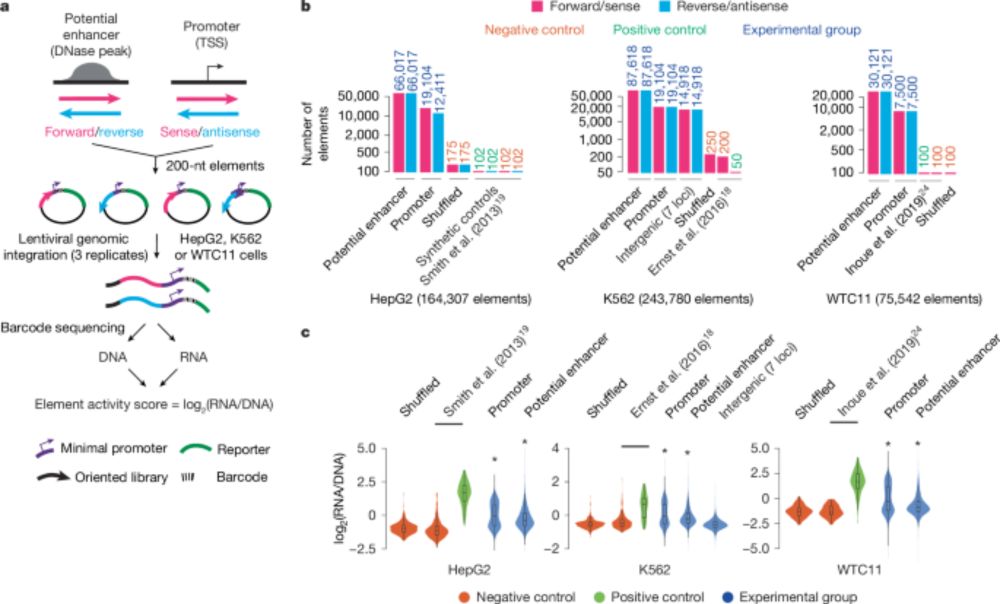
www.nature.com/articles/s41...
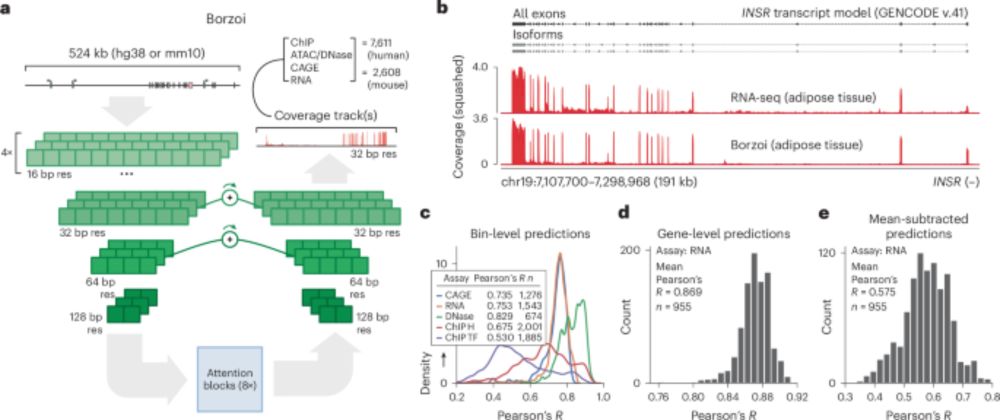
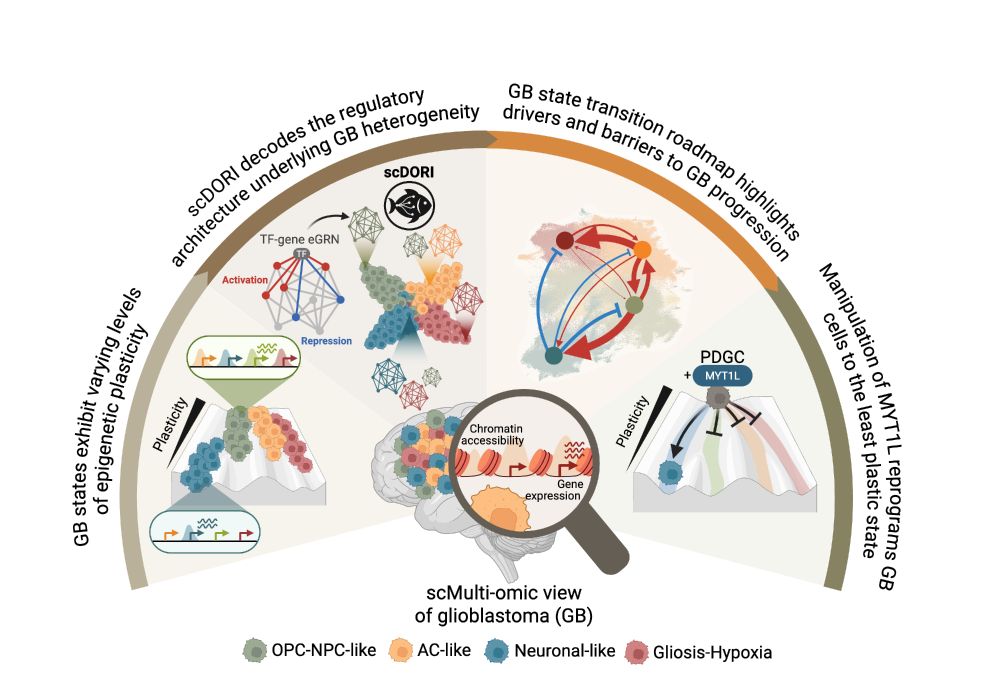
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

In a world of increasingly costly machine learning model deployments, ensuring accurate GPU operation timing is key to resource optimization. In this blog post, we explore best practices to achieve this in PyTorch.
www.speechmatics.com/company/arti...

In a world of increasingly costly machine learning model deployments, ensuring accurate GPU operation timing is key to resource optimization. In this blog post, we explore best practices to achieve this in PyTorch.
www.speechmatics.com/company/arti...
academic.oup.com/nar/advance-...

academic.oup.com/nar/advance-...

Part 1: go.bsky.app/5zgpZfg
Part 2: go.bsky.app/Q5sXc6E
Part 1: go.bsky.app/5zgpZfg
Part 2: go.bsky.app/Q5sXc6E


