
Happy to share the first results from our effort to reannotate Pseudomonas #phage proteins of unknown function, using structural bioinformatics.
📄 doi.org/10.1101/2025...
Instead of forcing structures into rigid classifications, we built AFragmenter. It uses AlphaFold PAE networks for a tuneable approach to domain parsing.
You control the granularity. 👇
🔗 doi.org/10.1093/bioi...

Instead of forcing structures into rigid classifications, we built AFragmenter. It uses AlphaFold PAE networks for a tuneable approach to domain parsing.
You control the granularity. 👇
🔗 doi.org/10.1093/bioi...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
My phage annotation tool, Phynteny, finally has a preprint and a brand new version powered by a cool AI transformer architecture and protein language models! #phagesky
www.biorxiv.org/content/10.1...

My phage annotation tool, Phynteny, finally has a preprint and a brand new version powered by a cool AI transformer architecture and protein language models! #phagesky
www.biorxiv.org/content/10.1...
Happy to share the first results from our effort to reannotate Pseudomonas #phage proteins of unknown function, using structural bioinformatics.
📄 doi.org/10.1101/2025...

Happy to share the first results from our effort to reannotate Pseudomonas #phage proteins of unknown function, using structural bioinformatics.
📄 doi.org/10.1101/2025...
💻🧬 #phagesky
linsalrob.github.io/PHROG_struct...
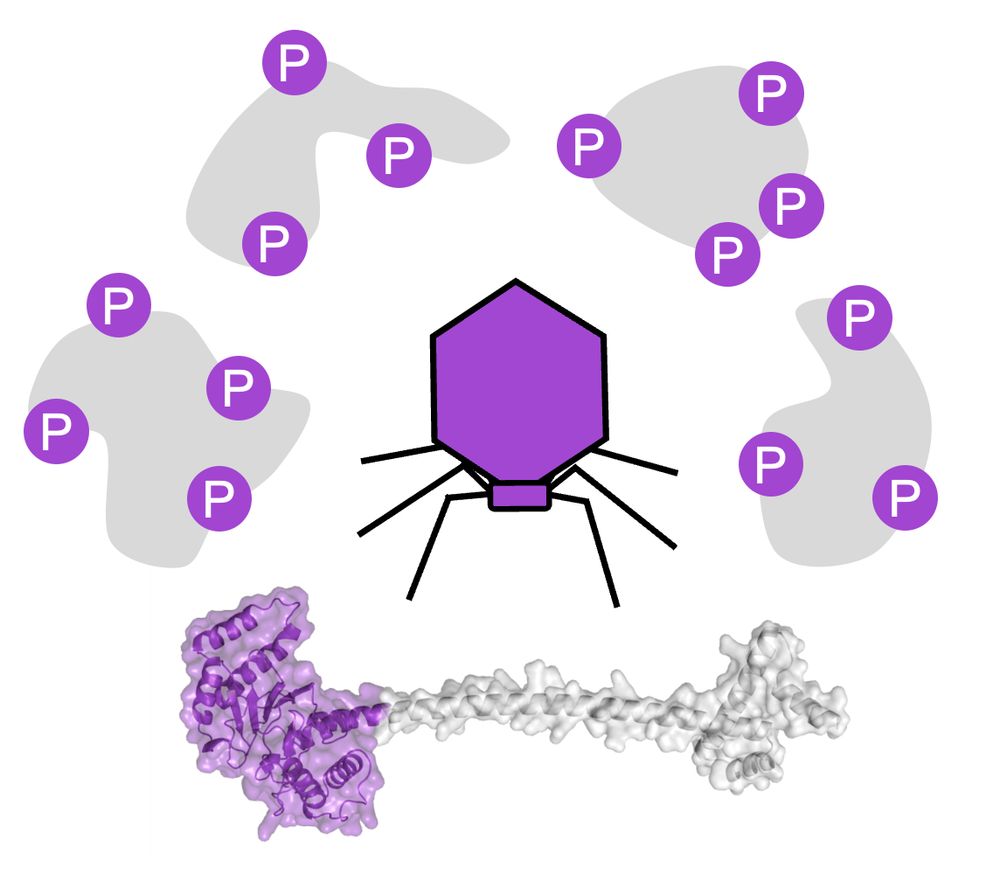
To celebrate, we have added handy new features to FLAMS web, incl. 3D visualization of hit proteins & PTM sites with known active/binding sites.
Come check it out @ tinyurl.com/flamsweb & let us know what you think!
To celebrate, we have added handy new features to FLAMS web, incl. 3D visualization of hit proteins & PTM sites with known active/binding sites.
Come check it out @ tinyurl.com/flamsweb & let us know what you think!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

