LOTs of takeaways from the Symposium (from biology to climate change)
🚨Here are a few important ones in my listicle below 👇
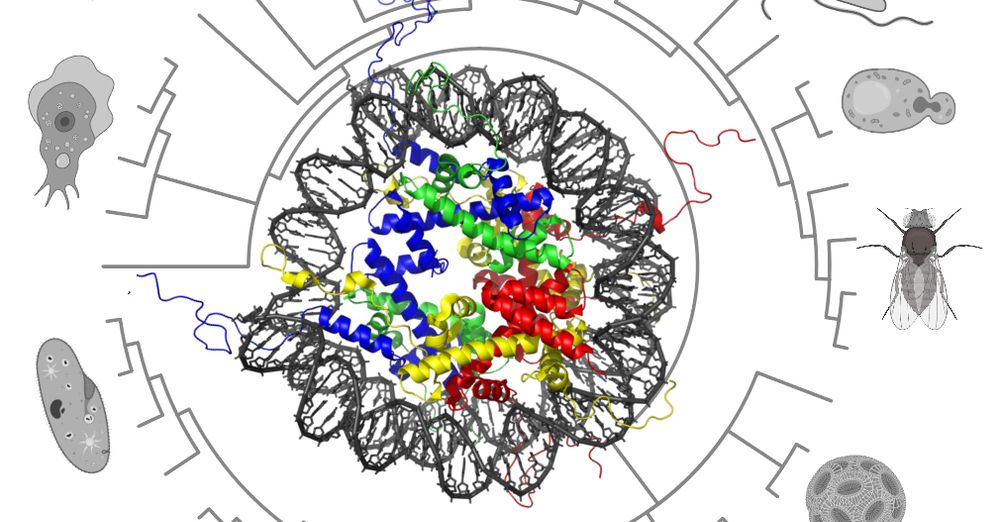
Multinucleate life really is everywhere...from deep sea forams to slime molds, intracellular parasites, and beyond. 🚀
We explore how scaling, ecology, & evolution intersect when many nuclei share a single cytoplasm.
doi.org/10.32942/X2M...
Multinucleate life really is everywhere...from deep sea forams to slime molds, intracellular parasites, and beyond. 🚀
We explore how scaling, ecology, & evolution intersect when many nuclei share a single cytoplasm.
doi.org/10.32942/X2M...
P.S. There are cartoons!
👉 tinyurl.com/4rz4ve6u

P.S. There are cartoons!
👉 tinyurl.com/4rz4ve6u
👉 github.com/JHelsen/point-centromere-detection
👉 github.com/JHelsen/point-centromere-detection
Discover it in our new Nature paper! We show centromeres transition gradually via a mix of drift, selection, and sex, reaching new states that still work with the kinetochore.
👉 doi.org/10.1038/s41586-025-09779-1

Discover it in our new Nature paper! We show centromeres transition gradually via a mix of drift, selection, and sex, reaching new states that still work with the kinetochore.
👉 doi.org/10.1038/s41586-025-09779-1
New research from EMBL and @Stanford shows how centromeres retain their function despite their rapid rate of change, and the evolutionary constraints that govern this process.
www.nature.com/articles/s41...

New research from EMBL and @Stanford shows how centromeres retain their function despite their rapid rate of change, and the evolutionary constraints that govern this process.
www.nature.com/articles/s41...
@gautamdey.bsky.social @helsenjana.bsky.social @gsherloc.bsky.social @kausthubh.bsky.social
@gautamdey.bsky.social @helsenjana.bsky.social @gsherloc.bsky.social @kausthubh.bsky.social
#protistsonsky 🧵
#protistsonsky 🧵
doi.org/10.32942/X2M...
We’d love your feedback while this goes through the peer review process!
#MicroEvoSky
#ProtistsonSky 🧪🌏


doi.org/10.32942/X2M...
We’d love your feedback while this goes through the peer review process!
#MicroEvoSky
#ProtistsonSky 🧪🌏
Is it sponges (panels A & B) or comb jellies (C & D) that root the animal tree of life?
For over 15 years, #phylogenomic studies have been divided.
We provide new evidence suggesting that...
🔗: www.science.org/doi/10.1126/...

Is it sponges (panels A & B) or comb jellies (C & D) that root the animal tree of life?
For over 15 years, #phylogenomic studies have been divided.
We provide new evidence suggesting that...
🔗: www.science.org/doi/10.1126/...
www.embl.org/about/info/e...
We have three positions in microbial genomics at EMBL-EBI, including one in my group. Please do apply, or if you know anyone that would be interested pass on to them
www.embl.org/about/info/e...
We have three positions in microbial genomics at EMBL-EBI, including one in my group. Please do apply, or if you know anyone that would be interested pass on to them
It grows as tangled filaments, divides unevenly, reshapes its own membranes… and even builds grappling hooks.
Meet Litorilinea aerophila — and here’s why it blew our minds.

It grows as tangled filaments, divides unevenly, reshapes its own membranes… and even builds grappling hooks.
Meet Litorilinea aerophila — and here’s why it blew our minds.
embl.wd103.myworkdayjobs.com/en-US/EMBL/j...
With @dudinlab.bsky.social
@embl.org @biology-unige.bsky.social @moorefound.bsky.social
embl.wd103.myworkdayjobs.com/en-US/EMBL/j...
With @dudinlab.bsky.social
@embl.org @biology-unige.bsky.social @moorefound.bsky.social
WitChi is:
✔ Fast
✔ Interpretable
✔ Tree- and model-free
✔ Benchmark-validated
Designed to fix compositional bias at phylogenomic scale.
With: @kassipan.bsky.social, @danieltamarit.bsky.social, @ettema.bsky.social
💻 github.com/stephkoest/w...
📄 www.biorxiv.org/content/10.1...
WitChi is:
✔ Fast
✔ Interpretable
✔ Tree- and model-free
✔ Benchmark-validated
Designed to fix compositional bias at phylogenomic scale.
With: @kassipan.bsky.social, @danieltamarit.bsky.social, @ettema.bsky.social
💻 github.com/stephkoest/w...
📄 www.biorxiv.org/content/10.1...
🧵 New preprint out!
WitChi: a fast, open-source Python tool to detect, quantify & prune compositional bias in MSAs.
Lightweight, tree-free, scalable to 5k+ taxa... so we applied it to the GTDB archaea MSA.
#ArchaeaSky #MEvoSky #MicroSky
🔗 doi.org/10.1101/2025...
💻 github.com/stephkoest/w...

🧵 New preprint out!
WitChi: a fast, open-source Python tool to detect, quantify & prune compositional bias in MSAs.
Lightweight, tree-free, scalable to 5k+ taxa... so we applied it to the GTDB archaea MSA.
#ArchaeaSky #MEvoSky #MicroSky
🔗 doi.org/10.1101/2025...
💻 github.com/stephkoest/w...
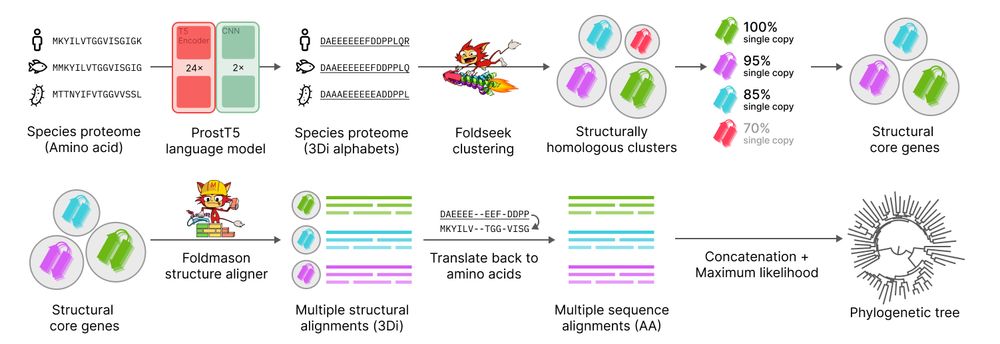
Unicore rapidly identifies structural single-copy core genes from input species proteomes for phylogenetic analysis. Powered by Foldseek and ProstT5, Unicore enables linear-scale structure-based phylogeny of any given set of taxa. 🧵1/n
📃 doi.org/10.1093/gbe/evaf109
Unicore rapidly identifies structural single-copy core genes from input species proteomes for phylogenetic analysis. Powered by Foldseek and ProstT5, Unicore enables linear-scale structure-based phylogeny of any given set of taxa. 🧵1/n
📃 doi.org/10.1093/gbe/evaf109
LOTs of takeaways from the Symposium (from biology to climate change)
🚨Here are a few important ones in my listicle below 👇
LOTs of takeaways from the Symposium (from biology to climate change)
🚨Here are a few important ones in my listicle below 👇
Learn more: www.embl.org/news/science...

Learn more: www.embl.org/news/science...


(Most credit to Minh Bui and @roblanfear.bsky.social and their labs)
ecoevorxiv.org/repository/v...
(Most credit to Minh Bui and @roblanfear.bsky.social and their labs)
ecoevorxiv.org/repository/v...
Massive thanks to @embl.org @events.embl.org and the organisers for putting this together. It was a real pleasure to interact with so many weird and wonderful creatures...
...and their model organisms (!!)

Massive thanks to @embl.org @events.embl.org and the organisers for putting this together. It was a real pleasure to interact with so many weird and wonderful creatures...
...and their model organisms (!!)
Sunset skies, science minds, and the EMBL DJ Club bringing the groove to the rooftop. Thanks to everyone who came out and made it such a special evening 🎧🌇 #EESWildModels
But the symposium isn't over yet! Time for the last talk of the day and poster prizes reveal 👀




Sunset skies, science minds, and the EMBL DJ Club bringing the groove to the rooftop. Thanks to everyone who came out and made it such a special evening 🎧🌇 #EESWildModels
But the symposium isn't over yet! Time for the last talk of the day and poster prizes reveal 👀
Thanks to our scientific organisers and to all who shared their science and energy. Congrats to our poster prize winners—exceptional work! 👏 #EESWildModels
Let’s keep exploring the unknown and see you next time!




Thanks to our scientific organisers and to all who shared their science and energy. Congrats to our poster prize winners—exceptional work! 👏 #EESWildModels
Let’s keep exploring the unknown and see you next time!