
We are uniting two research powerhouses.
For cutting-edge innovation that will boost our energy security, digital transformation, health and so much more.
Today is a good day for science, and for our EU-Switzerland partnership.

We are uniting two research powerhouses.
For cutting-edge innovation that will boost our energy security, digital transformation, health and so much more.
Today is a good day for science, and for our EU-Switzerland partnership.


Exciting topics ahead & we are looking forward to the #newPI talks by @julianeg.bsky.social (MPI) & Anupama Hemalatha (FMI).
www.ie-freiburg.mpg.de/grc

Exciting topics ahead & we are looking forward to the #newPI talks by @julianeg.bsky.social (MPI) & Anupama Hemalatha (FMI).
www.ie-freiburg.mpg.de/grc


📅 27.10. 16:30 – University of Bern, DCBP
📅 28.10. 15:00 – ETH Zurich, Hönggerberg
👉 More info: nccr-rna-and-disease.ch/education/nc...
@unibe.ch @ethz.ch @snf-fns.ch
📅 27.10. 16:30 – University of Bern, DCBP
📅 28.10. 15:00 – ETH Zurich, Hönggerberg
👉 More info: nccr-rna-and-disease.ch/education/nc...
@unibe.ch @ethz.ch @snf-fns.ch
scholar.google.com/citations?us...

scholar.google.com/citations?us...

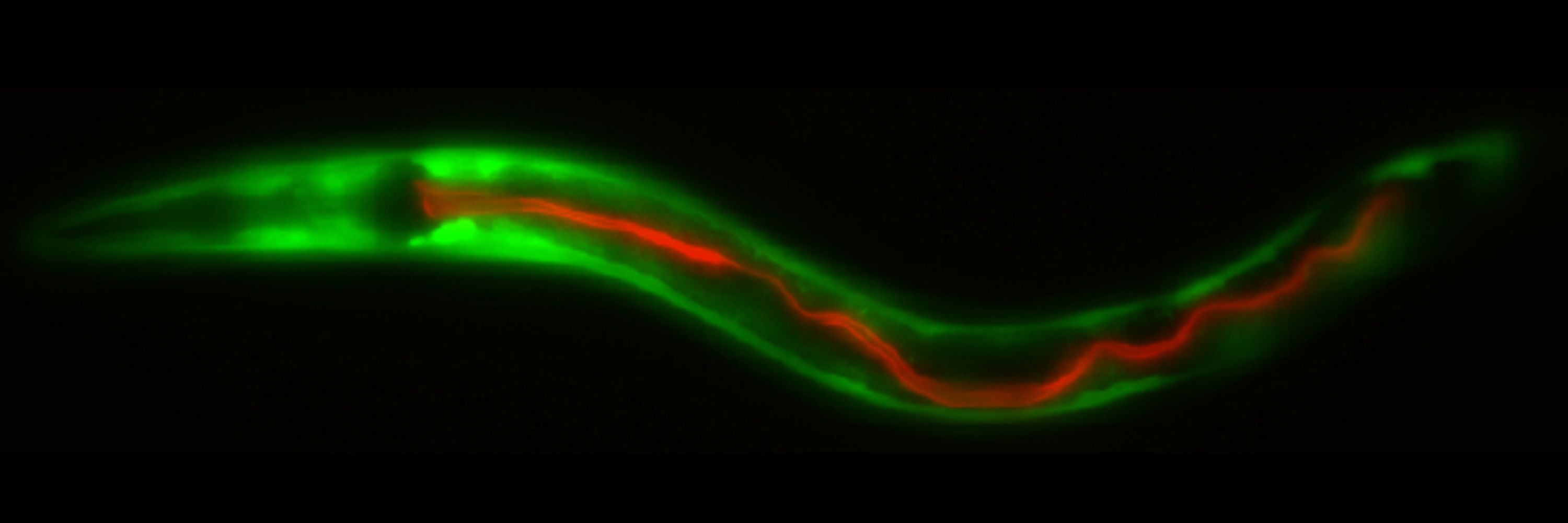
If you are interested in sensory biology and esp in cilia, thermosensation, or interoception, and would like to join an interactive & supportive group - please email.
Please RT 🙏

If you are interested in sensory biology and esp in cilia, thermosensation, or interoception, and would like to join an interactive & supportive group - please email.
Please RT 🙏








@unibe.ch @ethz.ch @snf-fns.ch

@unibe.ch @ethz.ch @snf-fns.ch

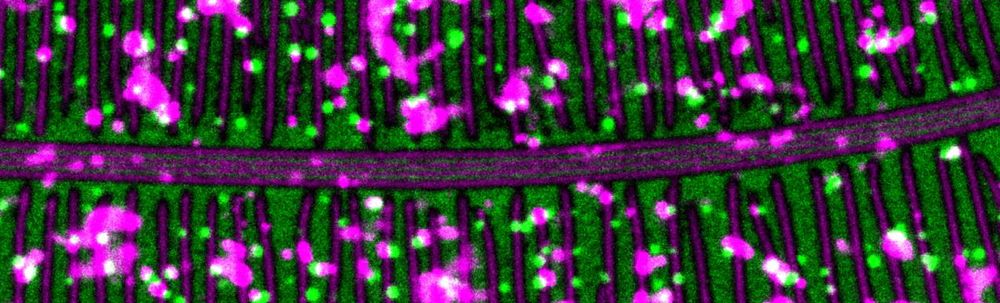
Innovative scientists in genome regulation, RNA metabolism, or protein homeostasis—especially using cutting-edge approaches—apply now at www.fmi.ch/education-ca...

Innovative scientists in genome regulation, RNA metabolism, or protein homeostasis—especially using cutting-edge approaches—apply now at www.fmi.ch/education-ca...
"A scheduler for rhythmic gene expression"
doi.org/10.1101/2025...

"Systematic identification of oscillatory gene expression in single cell types"
doi.org/10.1101/2025...

www.ie-freiburg.mpg.de/6057908/2025...

www.ie-freiburg.mpg.de/6057908/2025...
In project "InfoFate" we will study how cells use information in dynamical, neighborhood & mechanical signals to make decisions.
We'll have PhD and Postdoc positions available, please get in touch if interested!
Physicist Prof. David Brückner @davidbrueckner.bsky.social @biozentrum.unibas.ch @unibas.ch receives a prestigious #ERCStartingGrant to uncover how cells make life-shaping decisions. @erc.europa.eu
www.biozentrum.unibas.ch/news/detail/...

In project "InfoFate" we will study how cells use information in dynamical, neighborhood & mechanical signals to make decisions.
We'll have PhD and Postdoc positions available, please get in touch if interested!

