STORIES learns cell fate landscapes from spatial tramscripromics data profiled at several time points, thus allowing prediction of future cell states.
Led by Geert-Jan Huizing and Jules Samaran
www.nature.com/articles/s41...
@pasteur.fr

STORIES learns cell fate landscapes from spatial tramscripromics data profiled at several time points, thus allowing prediction of future cell states.
Led by Geert-Jan Huizing and Jules Samaran
www.nature.com/articles/s41...
@pasteur.fr
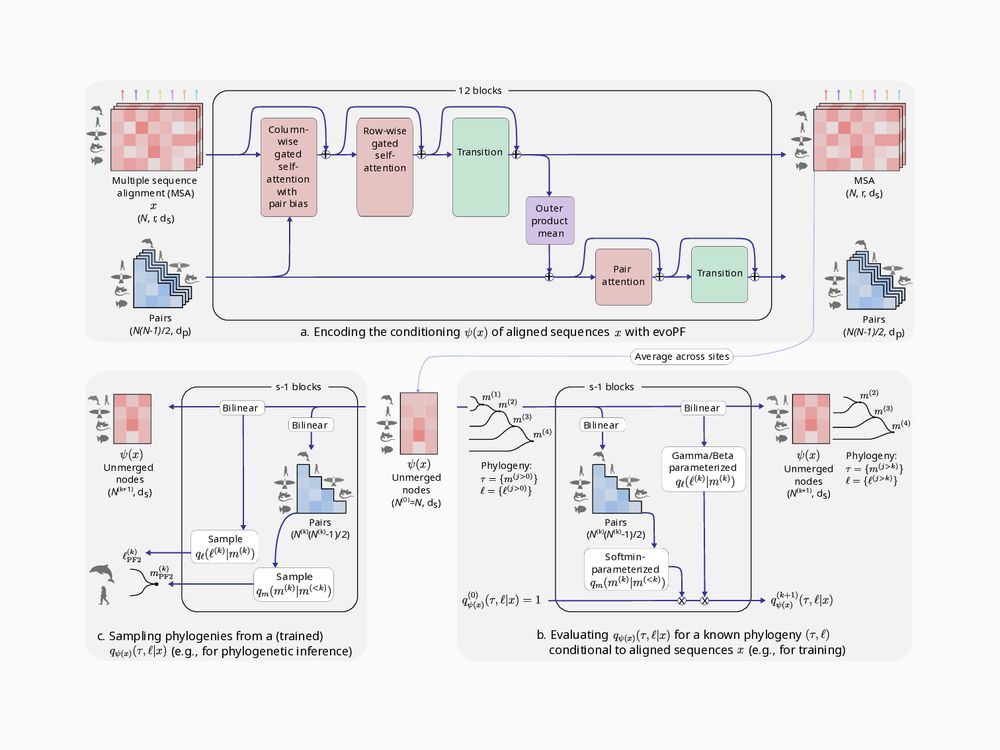
Phyloformer 2, a deep end-to-end phylogenetic reconstruction method: arxiv.org/abs/2510.12976
Using neural posterior estimation, it outperforms Phyloformer 1 and maximum-likelihood methods under simple and complex evolutionary models.
🧵1/17
The conference talks are online at legend2025.sciencesconf.org/data/book_le...
The conference talks are online at legend2025.sciencesconf.org/data/book_le...
www.nature.com/articles/d41...

www.nature.com/articles/d41...
I'm really looking forward to hearing these 21 exciting presentations (and additional 30 posters) next December.
If you want to attend too, registration is open until October 17th through legend2025.sciencesconf.org
![Exploring the space of self-reproducing RNA using generative models, Martin Weigt
Exploring the archaic introgression landscape of admixed populations through
joint ancestry inference, Jazeps Medina Tretmanis [et al.]
Predicting natural variation in the yeast phenotypic landscape with machine
learning, Sakshi Khaiwal [et al.]
Phylodynamic modeling with unsupervised Bayesian neural networks, Marino
Gabriele [et al.]
Likelihood-free inference of phylogenetic tree posterior distributions, Luc Blas-
sel [et al.]
Generative continuous time model reveals epistatic signatures in protein evolu-
tion, Barrat-Charlaix Pierre
Neural posterior estimation for high-dimensional genomic data from complex pop-
ulation genetic models, Jiseon Min [et al.]
A differentiable model for detecting diversifying selection directly from alignments
in large-scale bacterial datasets, Leonie Lorenz [et al.]
Detecting interspecific positive selection using transformers, Charlotte West [et al.]
Predicting Multiple Sequence Alignment Uncertainty via Machine Learning, Lucia
Martin-Fernandez [et al.]
Graph Neural Networks for Likelihood-Free Inference in Diversification Mod-
els, Amélie Leroy [et al.]
Popformer: learning general signatures of genetic variation and natural selection
with a self-supervised transformer, Leon Zong [et al.]](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:7x5c6ifrcznmg4mzhfuc2a6r/bafkreidur7bz43o2jhwr4gbt36awozawqthbkkfeszxjkxf4kvhyh7q6du@jpeg)
![PRIVET: PRIVacy metric based on Extreme value Theory, Antoine Szatkownik [et
al.]
Generative models for inferring the evolutionary history of the malaria vector
Anopheles gambiae, Amelia Eneli [et al.]
Language Models Outperform Supervised-Only Approaches for Conserved Ele-
ment Comprehension, Eyes Robson [et al.]
Identification and Classification of Orphan Genes, Spurious Orphan Genes, and
Conserved Genes from the human microbiome, Chen Chen
Neural Simulation-based inference of demography and selection, Francisco De
Borja Campuzano Jiménez [et al.]
Species Identification and aDNA Read Mapping Using k-mer Embeddings, Filip
Thor [et al.]
Contrastive Learning for Population Structure and Trait Prediction, Filip Thor [et
al.]
Protein and genomic language models chart a vast landscape of antiphage de-
fenses, Mordret Ernest
The Phylogenomics and Sparse Learning of Trait Innovations, Gaurav Diwan [et
al.]](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:7x5c6ifrcznmg4mzhfuc2a6r/bafkreig4akadduemzbit67bv7jnr7x2ircwiriykabg2imuf3phy6hhube@jpeg)
I'm really looking forward to hearing these 21 exciting presentations (and additional 30 posters) next December.
If you want to attend too, registration is open until October 17th through legend2025.sciencesconf.org




Logan now democratizes efficient access to the world’s most comprehensive genetics dataset. Free and open.
doi.org/10.1101/2024...

Logan now democratizes efficient access to the world’s most comprehensive genetics dataset. Free and open.
doi.org/10.1101/2024...
legend2025.sciencesconf.org
It will close on September 22nd (oral presentations) and October 1st (posters).
Send us your best work on Machine Learning for Evolutionary Genomics and come discuss it with us in the French Alps next December!
legend2025.sciencesconf.org
It will close on September 22nd (oral presentations) and October 1st (posters).
Send us your best work on Machine Learning for Evolutionary Genomics and come discuss it with us in the French Alps next December!
➡️ www.ins2i.cnrs.fr/fr/cnrsinfo/...
🤝 @lisnlab.bsky.social @cnrs-paris-saclay.bsky.social

➡️ www.ins2i.cnrs.fr/fr/cnrsinfo/...
🤝 @lisnlab.bsky.social @cnrs-paris-saclay.bsky.social
Our new abundance index, REINDEER2, is out!
It's cheap to build and update, offers tunable abundance precision at kmer level, and delivers very high query throughput.
Short thread!
www.biorxiv.org/content/10.1...
github.com/Yohan-Hernan...
Our new abundance index, REINDEER2, is out!
It's cheap to build and update, offers tunable abundance precision at kmer level, and delivers very high query throughput.
Short thread!
www.biorxiv.org/content/10.1...
github.com/Yohan-Hernan...
The 580€ include housing and all meals.
We will close on October 17th or when reaching 80 participants.
Mark your calendars and make sure your best work is ready next September when the call for abstracts opens 🙂
legend2025.sciencesconf.org

The 580€ include housing and all meals.
We will close on October 17th or when reaching 80 participants.
probgen2026.github.io
Please help spread the news.
probgen2026.github.io
Please help spread the news.
À lire sur le blog #FocusSciences🎯 du CNRS 👉 lejournal.cnrs.fr/nos-blogs/fo...

theses.hal.science/tel-00843343...
theses.hal.science/tel-00843343...
academic.oup.com/mbe/advance-...
The thread below provides a summary of our neural network for likelihood-free phylogenetic reconstruction.
academic.oup.com/mbe/advance-...
The thread below provides a summary of our neural network for likelihood-free phylogenetic reconstruction.




Mark your calendars and make sure your best work is ready next September when the call for abstracts opens 🙂
legend2025.sciencesconf.org

Mark your calendars and make sure your best work is ready next September when the call for abstracts opens 🙂
legend2025.sciencesconf.org
academic.oup.com/bioinformati...
Let's break it down:

academic.oup.com/bioinformati...
Let's break it down:

