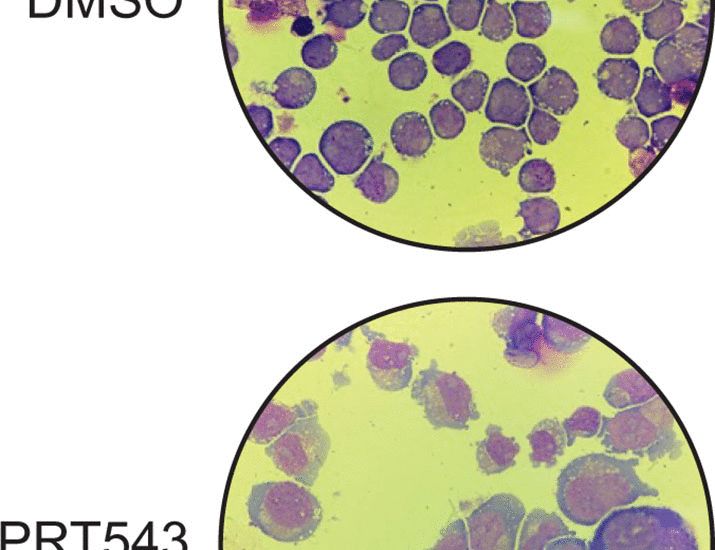
key point: 2/3rds of TF isos differ in properties like DNA binding & transcriptional activity
many are "negative regulators" & misexpressed in cancer
www.sciencedirect.com/science/arti...
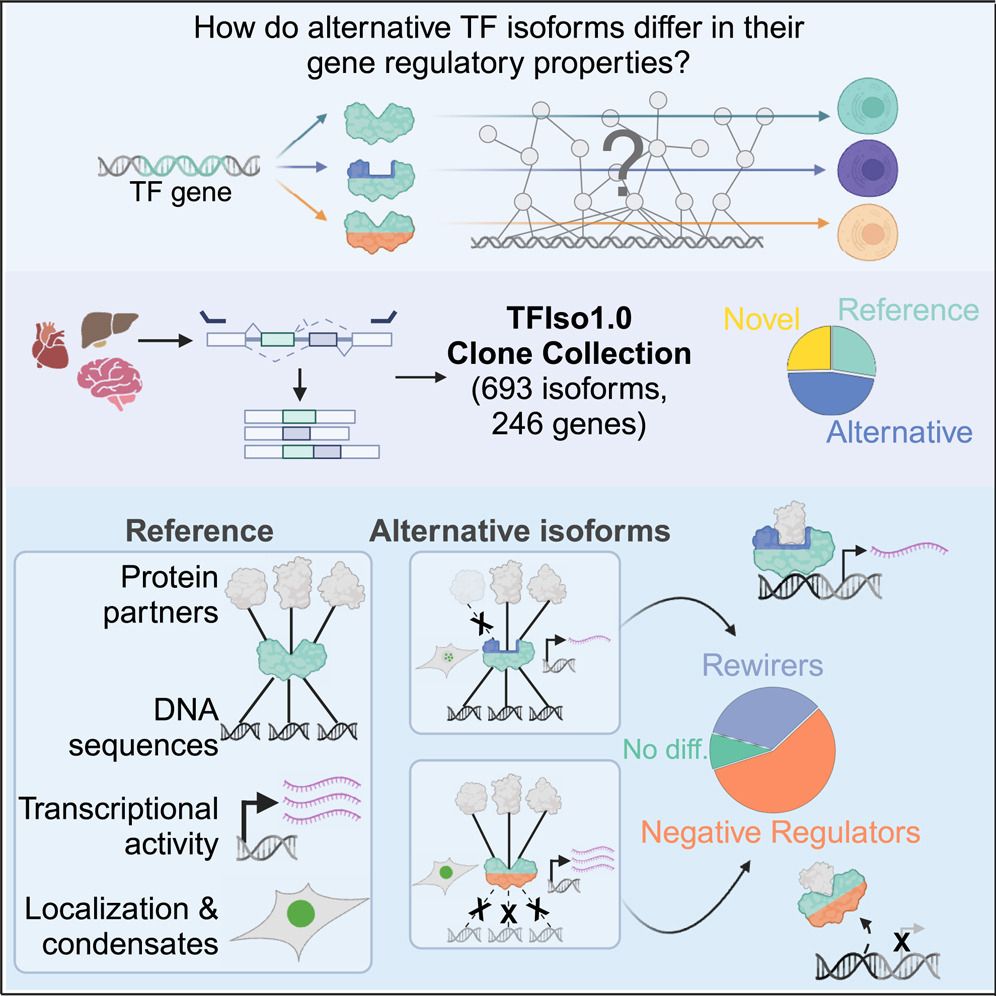
key point: 2/3rds of TF isos differ in properties like DNA binding & transcriptional activity
many are "negative regulators" & misexpressed in cancer
www.sciencedirect.com/science/arti...
doi.org/10.1038/s414...
#spatial #transcriptomics #scrnaseq #NMF
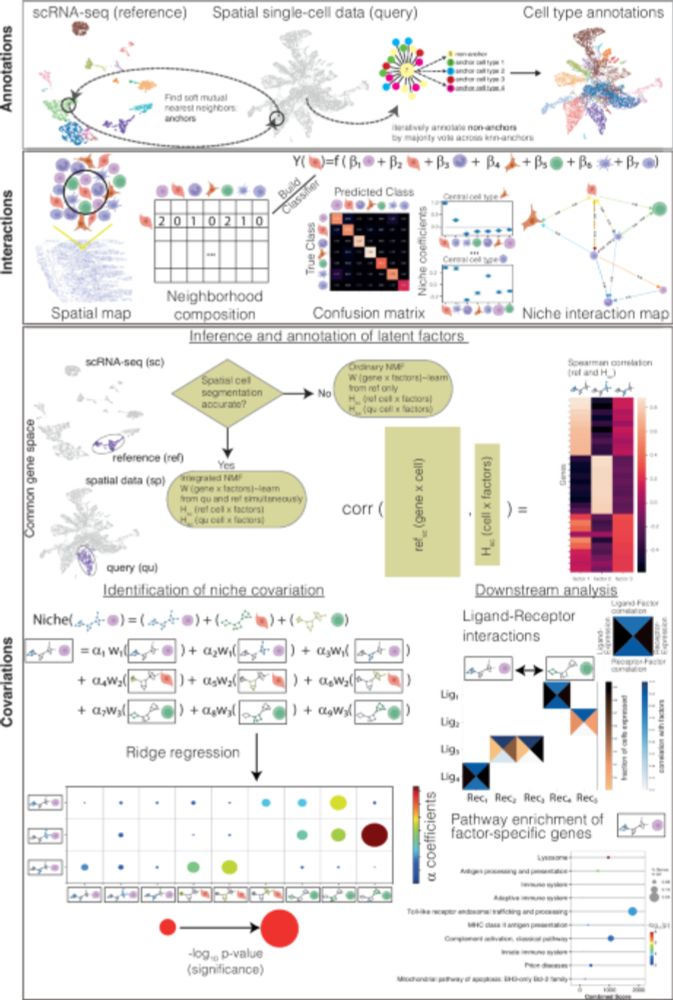
doi.org/10.1038/s414...
#spatial #transcriptomics #scrnaseq #NMF

Find out how it can serve you ⏬
🧵1/8
Find out how it can serve you ⏬
🧵1/8

