
@JLUGiessen, WGS bacteria, plasmids, software/pipeline developer, father of 2, husband, astrophotographer

@MicrobioSoc
#MGen
>661,000 bacterial genomes, uniformly characterized & annotated, enriched with metadata, accessible via a flexible search engine:
doi.org/10.1099/mgen...
👇 1/7

We celebrated 10 years of de.NBI with a birthday cake 🎂, welcomed new Associated Partners, and connected with 140+ participants. Stay tuned for more! #deNBI2025




We celebrated 10 years of de.NBI with a birthday cake 🎂, welcomed new Associated Partners, and connected with 140+ participants. Stay tuned for more! #deNBI2025
for the lasting support and funding enabling constant maintenance and development!
for the lasting support and funding enabling constant maintenance and development!
A huge shout out and thank you to all Bakta users, bug reporters, those sharing ideas and suggesting features...
...just the entire incredibly supporting binfie community!
Without you, Bakta wouldn't be the same.
Thank you!

A huge shout out and thank you to all Bakta users, bug reporters, those sharing ideas and suggesting features...
...just the entire incredibly supporting binfie community!
Without you, Bakta wouldn't be the same.
Thank you!
Please help us to further improve the annotation of bacterial genomes.
Please RT & share
(2/2)
Please help us to further improve the annotation of bacterial genomes.
Please RT & share
(2/2)
To further improve the functional annotation of "hypothetical" CDS, me and @gbouras13.bsky.social, we are looking for the worst Bakta-annotated bacterial genomes ;-)
(1/2)
To further improve the functional annotation of "hypothetical" CDS, me and @gbouras13.bsky.social, we are looking for the worst Bakta-annotated bacterial genomes ;-)
(1/2)
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
3 sessions full of microbial bioinformatics at JLU Giessen:
I: QC & QA, assembly
II: regional & functional annotation
III: comparative genomics
Info & registration: www.denbi.de/training-cou...
3 sessions full of microbial bioinformatics at JLU Giessen:
I: QC & QA, assembly
II: regional & functional annotation
III: comparative genomics
Info & registration: www.denbi.de/training-cou...
So, please use a proper encryption for sharing - if required.
So, please use a proper encryption for sharing - if required.
Of course, your data is handled with care automatically discarded after 30 days.
Just give it a try: bakta.computational.bio
(5/5)
Of course, your data is handled with care automatically discarded after 30 days.
Just give it a try: bakta.computational.bio
(5/5)
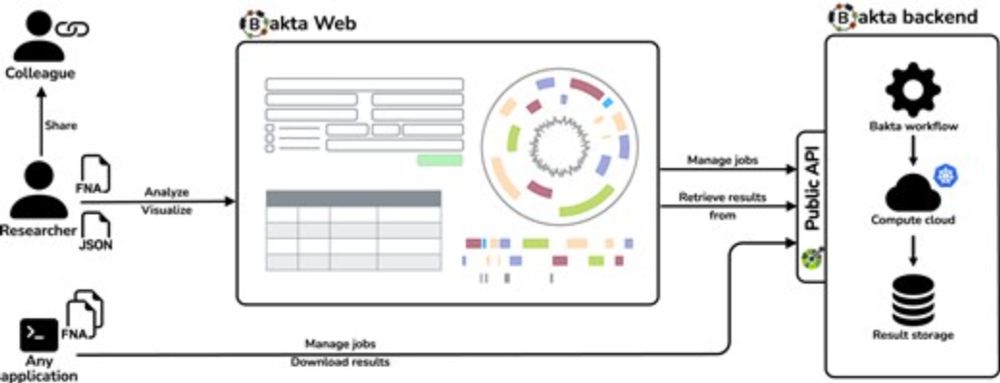
Of course, all results are also available for download in various standard file formats: GFF3, GenBank, EMBL, FASTA, TSV, TXT, JSON…
(4/5)
Of course, all results are also available for download in various standard file formats: GFF3, GenBank, EMBL, FASTA, TSV, TXT, JSON…
(4/5)
By this, Bakta Web dynamically scales out to meet temporal peak demands.
(3/5)
By this, Bakta Web dynamically scales out to meet temporal peak demands.
(3/5)
It provides interactive widgets supporting users with both, the provision of input data and sample/sequence metadata, and the interpretation of annotation results.
(2/5)
It provides interactive widgets supporting users with both, the provision of input data and sample/sequence metadata, and the interpretation of annotation results.
(2/5)
doi.org/10.1093/nar/...
Easy to use, no registration, fast, scalable, various visualizations, in sync with Bakta CLI:
bakta.computational.bio
(1/5)
doi.org/10.1093/nar/...
Easy to use, no registration, fast, scalable, various visualizations, in sync with Bakta CLI:
bakta.computational.bio
(1/5)
Hundreds of thousands, even millions?
All annotated, taxonomically classified, integrated with metadata.
Easily searchable, viewable, downloadable, in sync with #AllTheBacteria.
Then BakRep is for you! Poster P-CM-102 @vaam-microbes.bsky.social #VAAM25

Hundreds of thousands, even millions?
All annotated, taxonomically classified, integrated with metadata.
Easily searchable, viewable, downloadable, in sync with #AllTheBacteria.
Then BakRep is for you! Poster P-CM-102 @vaam-microbes.bsky.social #VAAM25
(6/6)
(6/6)
github.com/oschwengers/...
(5/6)
github.com/oschwengers/...
(5/6)
To mitigate negative download effects for our users, we therefore switched tarball compressions from gzip to xz.
Thus, compressed DB size actually decreased from 37GB to 30GB!
(4/6)
To mitigate negative download effects for our users, we therefore switched tarball compressions from gzip to xz.
Thus, compressed DB size actually decreased from 37GB to 30GB!
(4/6)
- COG: 2020 -> 2024
- Pfam: 36 -> 37.2
- RefSeq: r220 -> r228
- Rfam: 14.10 -> 15
- UniProtKB: 2023_05 -> 2025_01
Also, NCBI AMRFinderPlus and VFDB have been updated to the most recent versions.
(3/6)
- COG: 2020 -> 2024
- Pfam: 36 -> 37.2
- RefSeq: r220 -> r228
- Rfam: 14.10 -> 15
- UniProtKB: 2023_05 -> 2025_01
Also, NCBI AMRFinderPlus and VFDB have been updated to the most recent versions.
(3/6)
Numbers of IPS and PSC have grown by 22% and 13%.
(2/6)
Numbers of IPS and PSC have grown by 22% and 13%.
(2/6)
After a year, it was time for a Bakta database update - and it's a huge one:
- IPS: 330.9M
- PSC: 135.3M
- PSCC: 37M
doi.org/10.5281/zeno...
👇 1/6
After a year, it was time for a Bakta database update - and it's a huge one:
- IPS: 330.9M
- PSC: 135.3M
- PSCC: 37M
doi.org/10.5281/zeno...
👇 1/6
academic.oup.com/bioinformati...
academic.oup.com/bioinformati...

