
http://koo-lab.github.io

Excited to share SEAM: Systematic Explanation of Attribtuion-based Mechanisms. SEAM is an explainable AI method that dissects cis-regulatory mechanisms learned by seq2fun genomic deep learning models.
Led by @EESetiz
1/N 🧵👇



Excellent lineup of invited speakers across various scales of biology!
Deadline for abstract submission is coming up — Dec 2.
🔗 www.embl.org/about/info/c...
#EESAIBio @EMBLEvents

Excellent lineup of invited speakers across various scales of biology!
Deadline for abstract submission is coming up — Dec 2.
🔗 www.embl.org/about/info/c...
#EESAIBio @EMBLEvents
Excited to share SEAM: Systematic Explanation of Attribtuion-based Mechanisms. SEAM is an explainable AI method that dissects cis-regulatory mechanisms learned by seq2fun genomic deep learning models.
Led by @EESetiz
1/N 🧵👇

Excited to share SEAM: Systematic Explanation of Attribtuion-based Mechanisms. SEAM is an explainable AI method that dissects cis-regulatory mechanisms learned by seq2fun genomic deep learning models.
Led by @EESetiz
1/N 🧵👇
www.nobelprize.org/prizes/physi...
I have fond memories of my time in the Clarke lab, where I did my Honors Thesis on ultra low-field MRI w/ SQUIDs as an undergrad at UC Berkeley!

www.nobelprize.org/prizes/physi...
I have fond memories of my time in the Clarke lab, where I did my Honors Thesis on ultra low-field MRI w/ SQUIDs as an undergrad at UC Berkeley!
www.nature.com/articles/s41...
www.nature.com/articles/s41...


We have a great lineup of keynotes, contributed talks, and posters today and tomorrow
Schedule: mlcb.org/schedule
Join for free via livestream: m.youtube.com/@mlcbconf

We have a great lineup of keynotes, contributed talks, and posters today and tomorrow
Schedule: mlcb.org/schedule
Join for free via livestream: m.youtube.com/@mlcbconf
genomebiology.biomedcentral.com/articles/10....
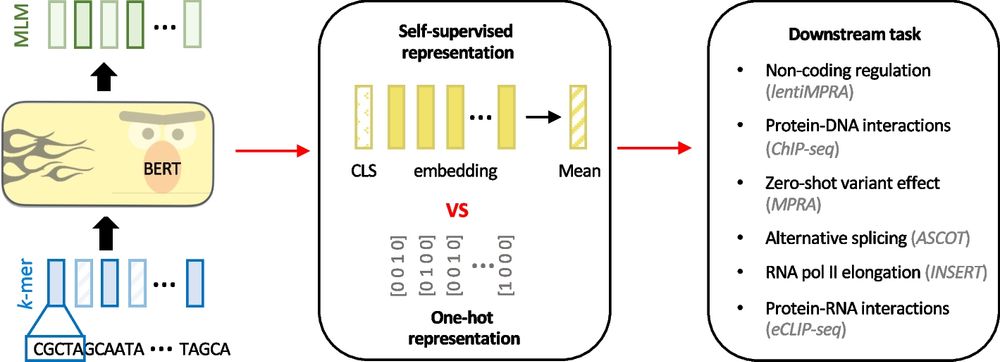
genomebiology.biomedcentral.com/articles/10....

genomebiology.biomedcentral.com/articles/10....
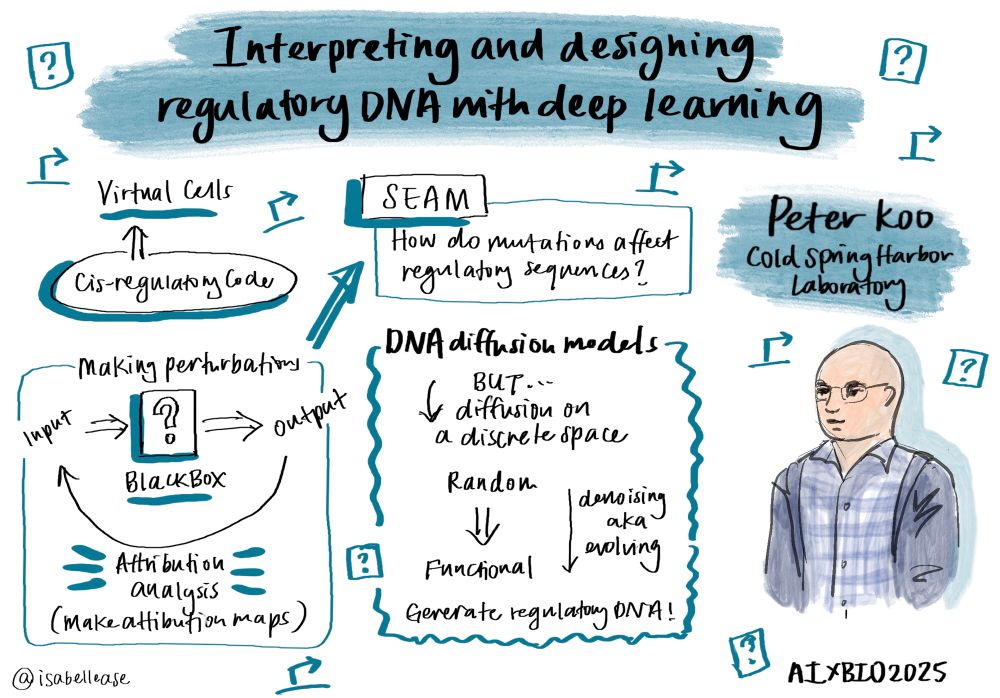

*Scaling* as the primary strategy with hopes of emergent properties is lazy.
Will the plan to fuse representations across mediocre (unimodal) foundation models work?!

*Scaling* as the primary strategy with hopes of emergent properties is lazy.
Will the plan to fuse representations across mediocre (unimodal) foundation models work?!
🧬 Paper: biorxiv.org/content/10.110…
(See thread below 👇) (1/n)
🧬 Paper: biorxiv.org/content/10.110…
(See thread below 👇) (1/n)
Submission deadline is June 1 for 2-page abstracts and 8-page papers (eligible for proceedings track).
Registration is now open! (Link below)
Please retweet!

Submission deadline is June 1 for 2-page abstracts and 8-page papers (eligible for proceedings track).
Registration is now open! (Link below)
Please retweet!

