Q-Immuno lab at UCL
@qimmuno.bsky.social
1.4K followers
660 following
24 posts
Quantitative immunology lab (PI: Andreas Tiffeau-Mayer)
https://qimmuno.com/
We use ideas from the physics of living systems and machine learning to understand human adaptive immunity in health and disease.
Posts
Media
Videos
Starter Packs
Reposted by Q-Immuno lab at UCL
Reposted by Q-Immuno lab at UCL
Reposted by Q-Immuno lab at UCL
Trevor Graham
@trevorgraham.bsky.social
· May 22

Data Scientist in Sutton | The Institute of Cancer Research
View details and apply for this Data Scientist vacancy in Sutton.
Salary : Salary range £39,805 to £49,023 (depending on the experience)
Reporting to: Professor Trevor Graham
...
jobs.icr.ac.uk
Reposted by Q-Immuno lab at UCL
Mahdad Noursadeghi
@mnoursad.bsky.social
· Apr 21

Evolution of T cell responses in the tuberculin skin test reveals generalisable Mtb-reactive T cell metaclones.
T cells contribute to immune protection and pathogenesis in tuberculosis, but measurements of polyclonal responses have failed to resolve correlates of outcome. We report the first temporal…
www.biorxiv.org
Reposted by Q-Immuno lab at UCL
Reposted by Q-Immuno lab at UCL
Reposted by Q-Immuno lab at UCL
Damon May
@the-dee-em.bsky.social
· Mar 19
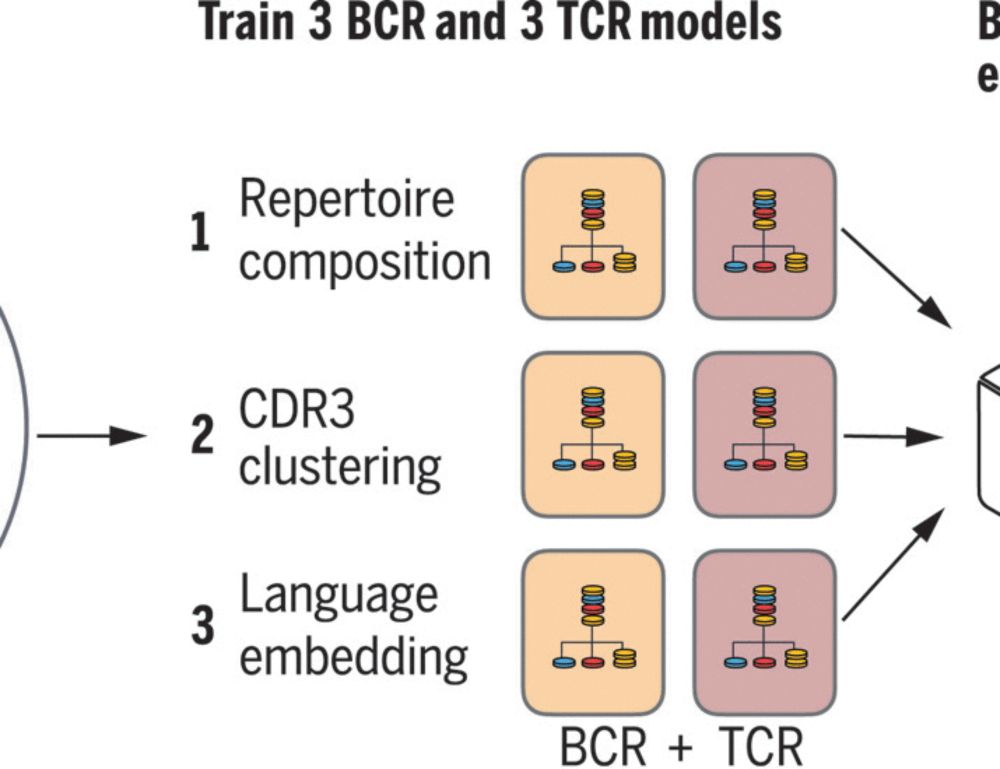
Disease diagnostics using machine learning of B cell and T cell receptor sequences
Clinical diagnosis typically incorporates physical examination, patient history, various laboratory tests, and imaging studies but makes limited use of the human immune system’s own record of antigen ...
www.science.org
Q-Immuno lab at UCL
@qimmuno.bsky.social
· Jan 17
Q-Immuno lab at UCL
@qimmuno.bsky.social
· Jan 17
Q-Immuno lab at UCL
@qimmuno.bsky.social
· Jan 17
Q-Immuno lab at UCL
@qimmuno.bsky.social
· Jan 17
Q-Immuno lab at UCL
@qimmuno.bsky.social
· Jan 17

Contrastive learning of T cell receptor representations
How should we train protein language models to predict protein function? This study
shows that pre-training using an autocontrastive loss greatly improves transfer learning
compared with masked-langua...
www.cell.com
Reposted by Q-Immuno lab at UCL






