

@aakaran31.bsky.social and @rivaselenarivas.bsky.social
link.springer.com/article/10.1...
July 05-17 2026
July 05-17 2026
benasque.org/2026rna/
Yes, Benasque is my profile pic. This #RNA meeting puts together for 2 weeks researchers in computational methods for structural RNA. 9th edition already! #RNAsky
benasque.org/2026rna/
Yes, Benasque is my profile pic. This #RNA meeting puts together for 2 weeks researchers in computational methods for structural RNA. 9th edition already! #RNAsky
@incarnatolab.bsky.social #rna #casprnasig #rnastructure

@incarnatolab.bsky.social #rna #casprnasig #rnastructure
github.com/marcellszi/r...
#RNA #RNAsky
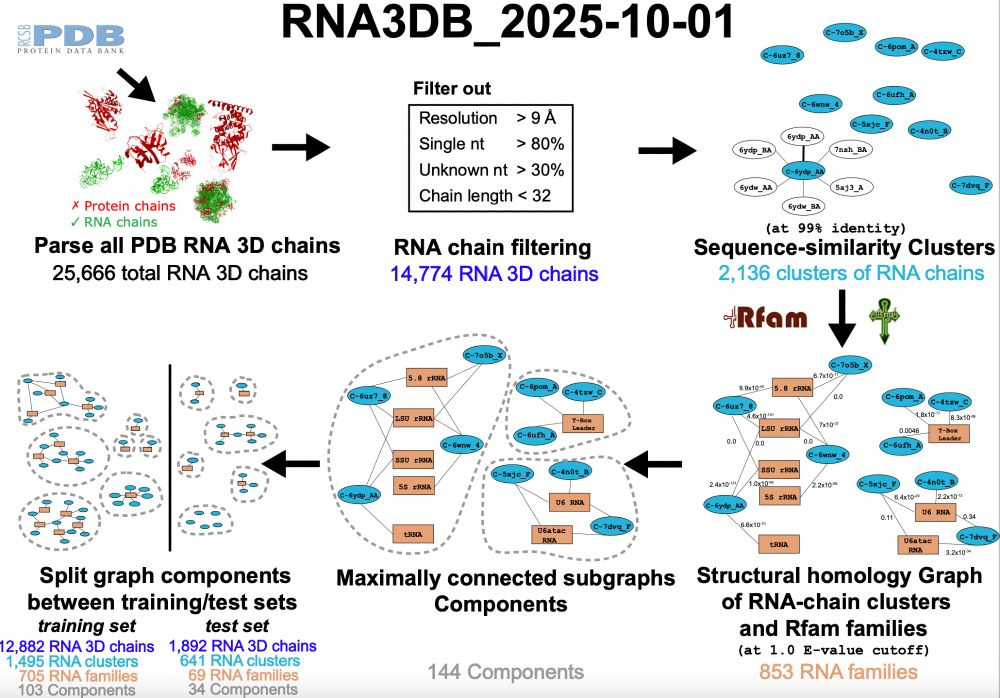
The database of all RNA chains in PDB arranged in structurally disimilar components, including Rfam annotation.
More chains (25,666), more independent components (144), more Rfam families represented (853).
github.com/marcellszi/r...
#RNA #RNAsky
The database of all RNA chains in PDB arranged in structurally disimilar components, including Rfam annotation.
More chains (25,666), more independent components (144), more Rfam families represented (853).
www.mcb.harvard.edu/department/n... @rivaselenarivas.bsky.social @rachellegaudet.bsky.social @cryptogenomicon.bsky.social @dulaclab.bsky.social @neurovenki.bsky.social @naoshigeuchida.bsky.social

www.mcb.harvard.edu/department/n... @rivaselenarivas.bsky.social @rachellegaudet.bsky.social @cryptogenomicon.bsky.social @dulaclab.bsky.social @neurovenki.bsky.social @naoshigeuchida.bsky.social
www.nature.com/articles/s41...

www.nature.com/articles/s41...

www.nature.com/articles/s41...
@aakaran31.bsky.social and @rivaselenarivas.bsky.social
link.springer.com/article/10.1...

@aakaran31.bsky.social and @rivaselenarivas.bsky.social
link.springer.com/article/10.1...

[Gift Link]
www.nytimes.com/2025/08/24/o...

[Gift Link]
www.nytimes.com/2025/08/24/o...
What does it take to learn the rules of RNA base pairing?
using standard deep-learning technics, got a simple answer:
don't need structures, nor alignments or many parameters
only a few RNA sequences and 21 parameters;
doi.org/10.1101/2025...
What does it take to learn the rules of RNA base pairing?
using standard deep-learning technics, got a simple answer:
don't need structures, nor alignments or many parameters
only a few RNA sequences and 21 parameters;
doi.org/10.1101/2025...

www.cambridgeday.com/2025/06/12/p...

www.cambridgeday.com/2025/06/12/p...
www.richmondscientific.com/how-a-discov...

www.richmondscientific.com/how-a-discov...
work lead (both computational and experimental) by then undergraduate, William Gao
work lead (both computational and experimental) by then undergraduate, William Gao

www.nytimes.com/interactive/...

www.nytimes.com/interactive/...
They have terminated ALL our funding, but we keep going.
Here is Dr Agata Kilar @amkilar.bsky.social presenting her work on conserved structural RNAs in nematodes.
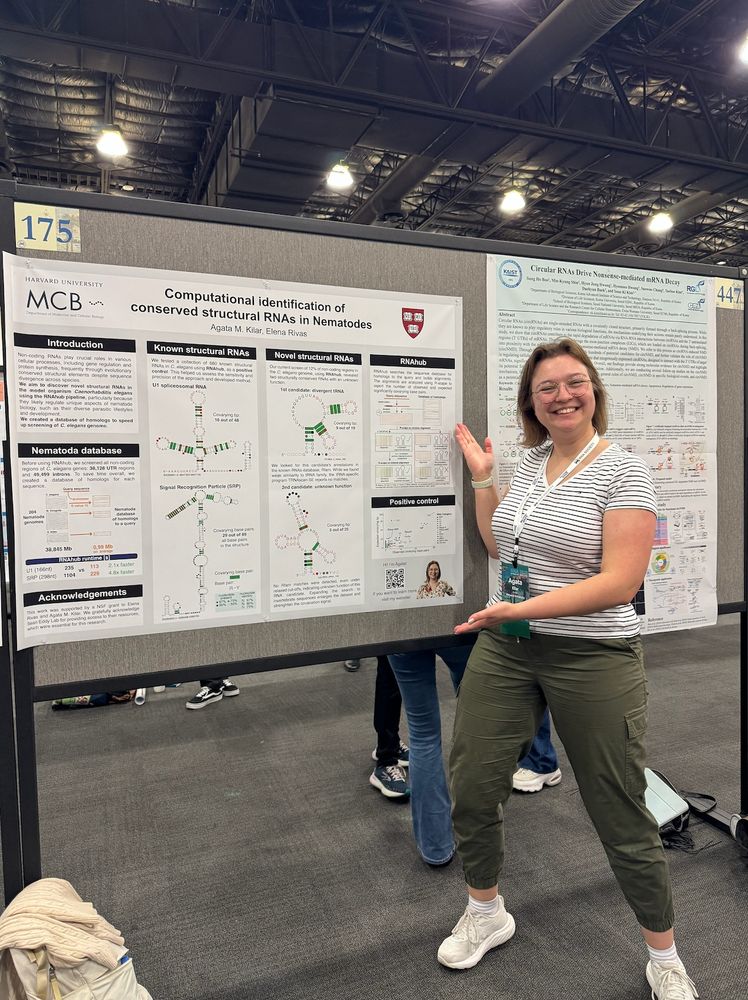
They have terminated ALL our funding, but we keep going.
Here is Dr Agata Kilar @amkilar.bsky.social presenting her work on conserved structural RNAs in nematodes.
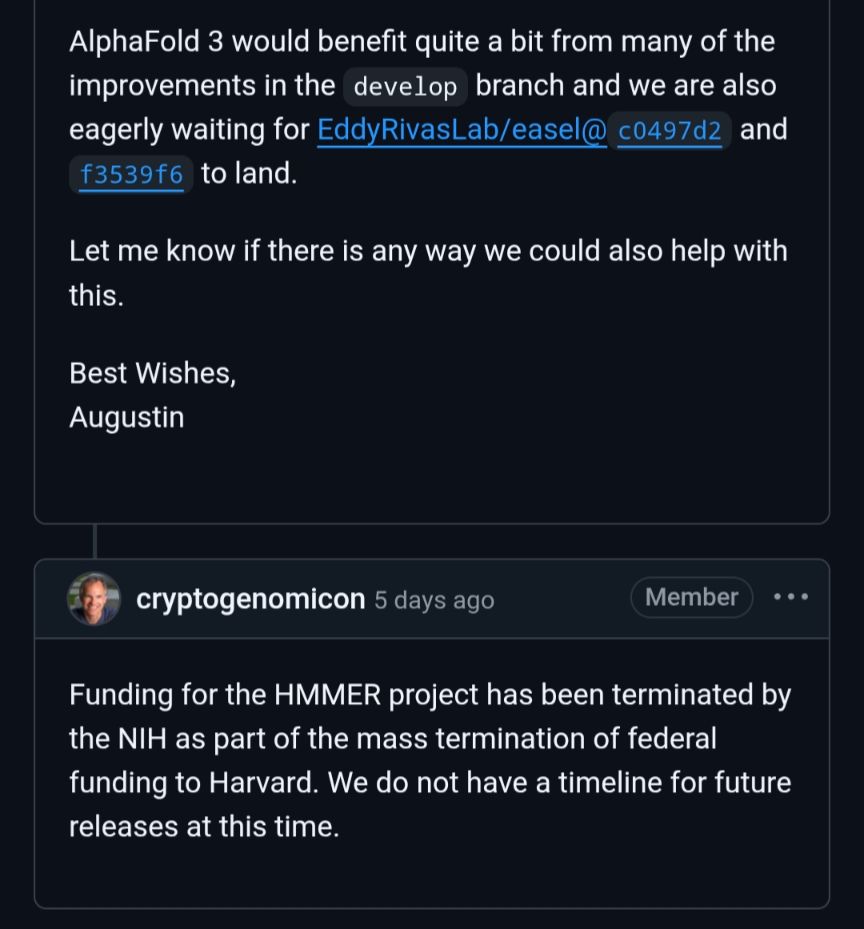
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...










