Elena Rivas
@rivaselenarivas.bsky.social
270 followers
350 following
29 posts
computational biologist. algorithms for genomics. worried about sustainability: from personal 2 country 2 planet. 🧪🧬| http://rivaslab.org
Posts
Media
Videos
Starter Packs
Reposted by Elena Rivas
Reposted by Elena Rivas
Laura Helmuth
@laurahelmuth.bsky.social
· Aug 30
jamelle
@jamellebouie.net
· Aug 29

RFK Jr.’s damage to the CDC is ‘past the point of no return,’ Dr. Demetre Daskalakis warns
“The CDC you knew is over,” the infectious diseases doctor told The Advocate. “Unless someone takes radical action, there is nothing there that can be salvaged.”
www.advocate.com
Elena Rivas
@rivaselenarivas.bsky.social
· Aug 28
Reposted by Elena Rivas
Reposted by Elena Rivas
Reposted by Elena Rivas
John Hawkinson
@johnhawkinson.bsky.social
· Jun 12

Petrova released on bail from federal detention, walks out of courthouse; indictment still possible - Cambridge Day
Harvard researcher Kseniia Petrova was released on bail after a 10-minute hearing in her criminal smuggling case – of frog embryos meant for scientific use.
www.cambridgeday.com
Reposted by Elena Rivas
Reposted by Elena Rivas
Elena Rivas
@rivaselenarivas.bsky.social
· May 29

All-at-once RNA folding with 3D motif prediction framed by evolutionary information
Structural RNAs exhibit a vast array of recurrent short 3D elements found in loop regions involving non-Watson-Crick interactions that help arrange canonical double helices into tertiary structures. W...
doi.org
Reposted by Elena Rivas
Reposted by Elena Rivas
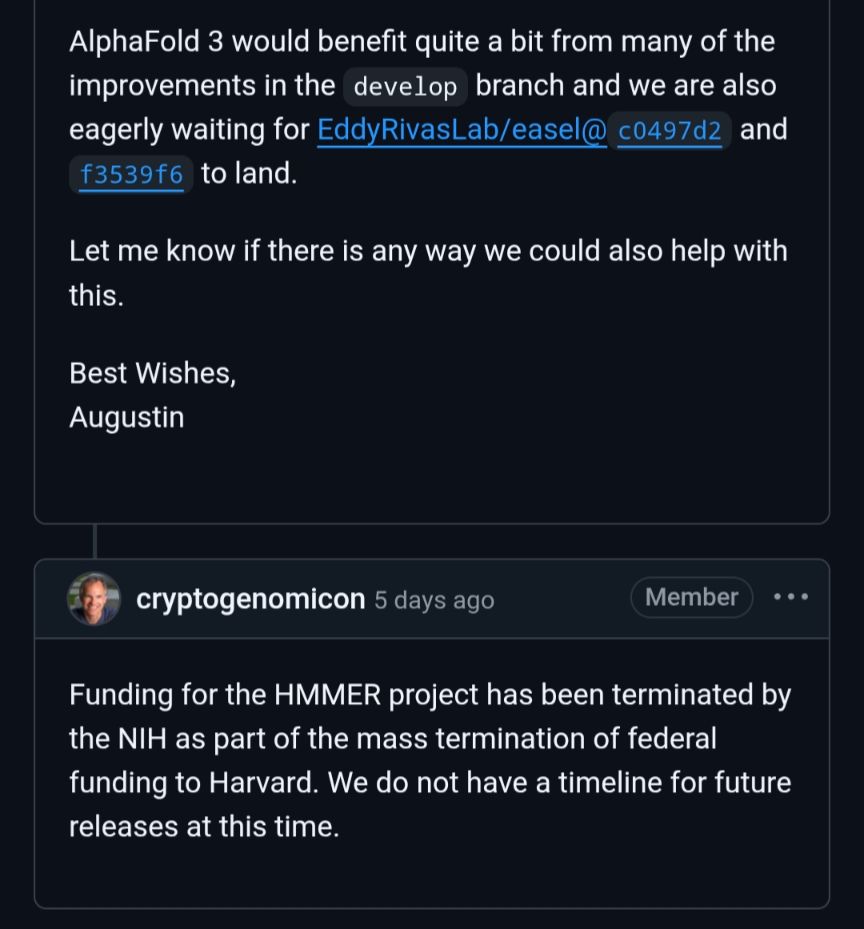
Sean Eddy
@cryptogenomicon.bsky.social
· May 26
Reposted by Elena Rivas
Liana Merk
@bacteriyay.bsky.social
· May 24

Prevalence of Group II Introns in Phage Genomes
Although bacteriophage genomes are under strong selective pressure for high coding density, they are still frequently invaded by mobile genetic elements (MGEs). Group II introns are MGEs that reduce h...
www.biorxiv.org
Reposted by Elena Rivas
Jon Levy
@jonlevybu.bsky.social
· May 15
Jacquelyn Gill
@jacquelyngill.bsky.social
· May 14

Few support punitive funding cuts to colleges and universities - AP-NORC
Most of the public believes that colleges and universities make significant contributions to scientific and technological advancement and think their federal research funding should be maintained.
apnorc.org